Relationship to other mycobacterial pathogens
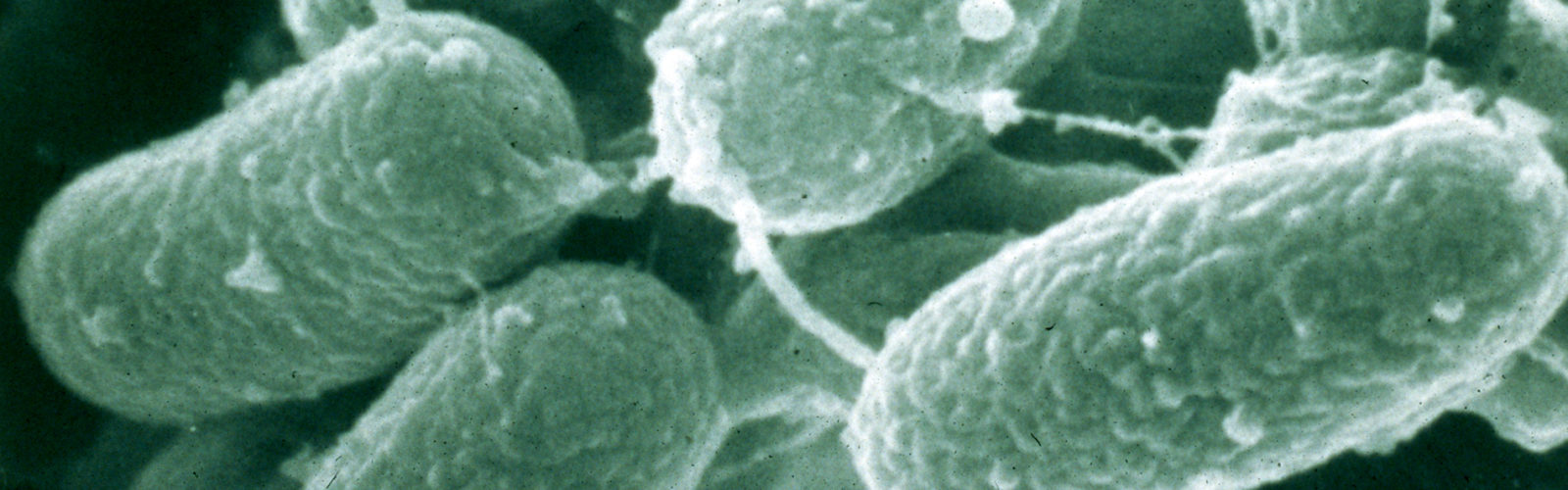
 Mycobacterium paratuberculosis was reclassified as a subspecies of the Mycobacterium Avium Complex (MAC) and is now correctly called Mycobacterium avium subspecies paratuberculosis, commonly abbreviated MAP. This was first proposed by Dr. Marie Thorel in 1990. A partial phylogenetic tree of the relationship among the slow-growing pathogenic mycobacteria based on 16S rDNA sequencing is shown above.
Mycobacterium paratuberculosis was reclassified as a subspecies of the Mycobacterium Avium Complex (MAC) and is now correctly called Mycobacterium avium subspecies paratuberculosis, commonly abbreviated MAP. This was first proposed by Dr. Marie Thorel in 1990. A partial phylogenetic tree of the relationship among the slow-growing pathogenic mycobacteria based on 16S rDNA sequencing is shown above.
Genetic studies reveal that MAP is a clone of closely related organisms derived from the parent Mycobacterium avium subspecies hominissuis. Despite genetic similarity, many observable (phenotypic) characteristics distinguish MAP from other members of the MAC. Some of MAP’s unique characteristics are:
- MAP grows much more slowly (growth rate of mycobacteria is associated with pathogenicity; slower-growth related to greater pathogenicity).
- MAP requires an iron-transport compound known as mycobactin for in vitro growth (mycobactin synthesis allows other mycobacteria to replicate outside their host).
- MAP primarily infects ruminant mammals instead of birds.
- MAP is more virulent, capable of infecting and causing disease in animals with a fully competent immune system. Most other members of the MAC are considered opportunistic pathogens attacking individuals with a compromised immune system.
- Young animals are more susceptible to MAP infection than adults for reasons that are not known. Some suggest that neonatal ruminants do not have a fully developed cellular immune system.
- MAP infections primarily target the intestinal tract.
MAP and other members of the Mycobacterium Avium Complex (MAC) are less closely related genetically to pathogenic mycobacteria in the TB complex: Mycobacterium tuberculosis, the cause of tuberculosis in humans, and Mycobacterium bovis, the cause of tuberculosis in cattle and other animals. MAP also is not closely related to the cause of leprosy in humans, Mycobacterium leprae. MAP, and the disease it causes in animals (Johne’s disease), does however share certain biological characteristics in common with these mycobacterial pathogens. Scientists often draw parallels among these mycobacterial organisms to try and understand their basic mechanisms for causing disease.
Environmental distribution
MAP bacteria are not thought to be free-living (able to grow and multiply) in the environment. Because of its inability to produce mycobactin (a chemical needed to transport iron), MAP can grow only inside animal cells where it “steals” iron from its host’s cells, most often immune cells called macrophages. Thus, it is an obligate parasitic pathogen of mammals. This means infected animals are the only place in nature where growth and multiplication of MAP can occur. If found in soil or water samples, it can be assumed that MAP is simply persisting in those places (not multiplying) after being deposited there through fecal contamination from an infected animal.
MAP is extremely hardy and may remain in contaminated soil and water for a year or more. From an anthropomorphic perspective MAP, compensates for its inability gather iron and replicate in the environment (due to lack of mycobactin) by producing an extra tough outer coat enabling prolonged survival while it waits to be eaten by a susceptible host, enter host macrophages, and resume replication.
The environmental distribution of MAP is markedly different from that most other members of the Mycobacterium Avium Complex (MAC), in particular the ubiquitous M. avium subspecies hominissuis (MAH). MAH is commonly found in lakes, streams and domestic water supplies. Certain acidic soil types, notably peat bogs, contain higher than average numbers of MAH. MAP only occurs in environments contaminated by feces of infected animals. A tenuous association between the occurrence of Johne’s disease and geographical regions with acidic soils has been reported (see discussion under “survival in soil” on this website). The strength of this association and the biological basis of this association remain to be determined.
Host range

MAP has a broad host range. Ruminants (animals with a four-chambered stomach that chew their cud) are the type of animal most commonly infected. These include: cattle, sheep, goats, deer, elk, antelope, bison, camels, llamas, and alpacas (these latter three species are technically called pseudo-ruminants as they have a three-chambered stomach). There are also infrequent reports of MAP infections (but not subsequent disease) in non-ruminant species such as carnivorous or omnivorous predators (weasels, , fox, stoat, crows, coyotes, raccoons, etc.). Infection in prey animals (mice, rabbits, vole, rat, sparrows, etc.) have been noted; disease is rare in these species except for some cases of rabbits (Greig et al. 1999).
See “Zoonotic Potential” for a discussion of MAP in humans. If you wish to focus on particular species, select it at the left of your screen. You will then be able to choose the particular Johne’s disease topic in which you are interested.
Virulence factors
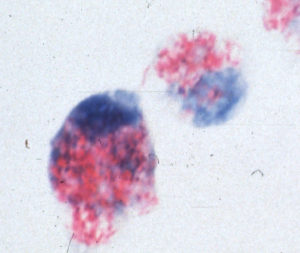 Like other mycobacteria, MAP has the capacity to thrive inside white blood cells known as macrophages. As part of the immune system, macrophages are capable of destroying a wide variety of bacterial pathogens. Mycobacteria, however, are one of the few types of bacteria that not only can survive the antibacterial effects of macrophages, but actually grow and multiply inside them. Bacteria able to avoid being killed and instead replicate inside macrophages and cause disease are referred to as facultative intracellular bacterial pathogens. This picture shows MAP (red) inside macrophages (blue).
Like other mycobacteria, MAP has the capacity to thrive inside white blood cells known as macrophages. As part of the immune system, macrophages are capable of destroying a wide variety of bacterial pathogens. Mycobacteria, however, are one of the few types of bacteria that not only can survive the antibacterial effects of macrophages, but actually grow and multiply inside them. Bacteria able to avoid being killed and instead replicate inside macrophages and cause disease are referred to as facultative intracellular bacterial pathogens. This picture shows MAP (red) inside macrophages (blue).
Considerable research has been done to try and understand how mycobacteria flourish in what is thought to be the hostile intracellular environment of macrophages. Science is closing in on understanding the specific mechanisms for the very effective mycobacterial strategy. In general terms, three properties of mycobacteria explain their resistance to being killed by macrophages: 1) the chemically unique mycobacterial cell wall that is resistant to destruction or penetration, 2) secretion of factors by mycobacteria that avoid or neutralize the antibacterial chemicals produced inside macrophages, and 3) secretion of chemicals that suppress or modulate the host’s immune response. For detailed information the reader should obtain the references listed at the end of this section on the biology of MAP.
Survival and multiplication in the host animal is a prerequisite to causing disease. As with mechanisms of intracellular survival, mechanisms by which mycobacteria cause disease are also not well understood. Pathology due to mycobacterial infections results in part from the direct action of toxic chemical components of the mycobacterial cell wall. The host animal’s immune response to the presence of MAP also contributes to the pathology and dysfunction resulting from the infection.
Colonial morphology
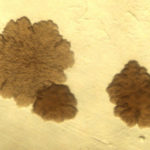 The
The size, color, and texture of a colony of MAP is dependent in part on the type of bacteriologic medium on which it is cultivated. On Herrold’s egg yolk agar medium, one of the most commonly used culture mediums in veterinary diagnostic laboratories, the colonies appear small, somewhat rough and off-white to yellow in color. Pigmented (yellow) strains have been reported in sheep. On Middlebrook agar medium without Tween 80 (a detergent that improves the growth rate) the colonies are very rough in appearance and resemble those of M. tuberculosis. With addition of Tween 80 the growth rate of MAP increases and it’s colonial morphology becomes smooth and domed resembling that of M. avium subsp. avium (MAA).
size, color, and texture of a colony of MAP is dependent in part on the type of bacteriologic medium on which it is cultivated. On Herrold’s egg yolk agar medium, one of the most commonly used culture mediums in veterinary diagnostic laboratories, the colonies appear small, somewhat rough and off-white to yellow in color. Pigmented (yellow) strains have been reported in sheep. On Middlebrook agar medium without Tween 80 (a detergent that improves the growth rate) the colonies are very rough in appearance and resemble those of M. tuberculosis. With addition of Tween 80 the growth rate of MAP increases and it’s colonial morphology becomes smooth and domed resembling that of M. avium subsp. avium (MAA).
Cellular morphology
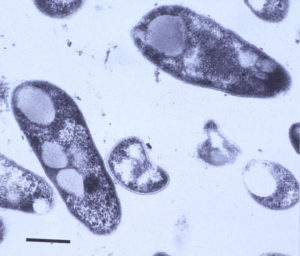 MAP is a small (0.5 x 1.5 micron) rod-shaped bacterium, roughly the size of the common intestinal bacterium called E. coli, that grows in clumps of 10 to 100 bacterial cells. It can be seen using a light microscope with 40x or greater power objectives. When stained by the gram stain, it is blue and so called gram-positive. When stained by acid-fast stains like the Ziehl-Neelsen or Kinyoun’s stain, MAP stains red and so is called acid-fast positive. Transmission electron microscopy reveals the waxy rough cell wall of M. paratuberculosis with its trilaminar structure. The intracellular vacuoles or inclusions common to mycobacteria can also be seen.
MAP is a small (0.5 x 1.5 micron) rod-shaped bacterium, roughly the size of the common intestinal bacterium called E. coli, that grows in clumps of 10 to 100 bacterial cells. It can be seen using a light microscope with 40x or greater power objectives. When stained by the gram stain, it is blue and so called gram-positive. When stained by acid-fast stains like the Ziehl-Neelsen or Kinyoun’s stain, MAP stains red and so is called acid-fast positive. Transmission electron microscopy reveals the waxy rough cell wall of M. paratuberculosis with its trilaminar structure. The intracellular vacuoles or inclusions common to mycobacteria can also be seen.
Cell wall chemistry
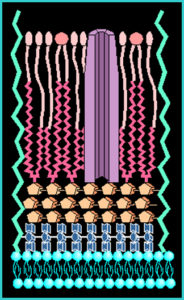 The cell wall of mycobacteria is composed of a thick waxy mixture of lipids and polysaccharides. The cell wall of MAP, although not well studied, seems similar in most respects to that of other mycobacteria. One feature is notable, however. While most strains of M. avium produce a surface glycolipid that allows strains to be serotyped (i.e., distinguished using antibodies specific for each glycolipid subtype). MAP strains lack such glycolipid antigens on their surface.
The cell wall of mycobacteria is composed of a thick waxy mixture of lipids and polysaccharides. The cell wall of MAP, although not well studied, seems similar in most respects to that of other mycobacteria. One feature is notable, however. While most strains of M. avium produce a surface glycolipid that allows strains to be serotyped (i.e., distinguished using antibodies specific for each glycolipid subtype). MAP strains lack such glycolipid antigens on their surface.
See Wikipedia for a more comprehensive discussion of mycobacteria.
Biochemical characteristics
Biochemical tests used to distinguish among other species of mycobacteria are not used to identify MAP. The tests are taxing to perform due to the extremely slow growth rate of the organism, and test results vary among strains of MAP. However, in a seminal publication, Dr. Marie-Françoise Thorel (photo) and colleagues described biochemical tests and numerical taxonomy methods to differentiate among subtypes of MAP-like bacteria. This work led to the adoption of the new name Mycobacterium avium subspecies paratuberculosis (MAP).
 Dr. Thorel passed away in 2017 after a highly productive career. She retired in 2003 from her post at the Maisons-Alfort Laboratory for Animal Health after a long career in the paratuberculosis research field, being one of the participants in the first International Colloquium on Paratuberculosis (ICP) held in Ames, Iowa in 1983. She was the organizer of the Second Colloquium in Paris in 1988 from which the International Association for Paratuberculosis (IAP) started. She a co-founder of the IAP and member of the Executive Board representing France for several years. Photo taken by M.T. Collins at one of the ICP meetings.
Dr. Thorel passed away in 2017 after a highly productive career. She retired in 2003 from her post at the Maisons-Alfort Laboratory for Animal Health after a long career in the paratuberculosis research field, being one of the participants in the first International Colloquium on Paratuberculosis (ICP) held in Ames, Iowa in 1983. She was the organizer of the Second Colloquium in Paris in 1988 from which the International Association for Paratuberculosis (IAP) started. She a co-founder of the IAP and member of the Executive Board representing France for several years. Photo taken by M.T. Collins at one of the ICP meetings.
Molecular genetics
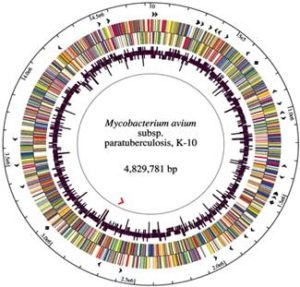 The DNA of MAP is >99% identical with that of M. avium, hence the change in naming as described above. The K-10 strain of MAP was fully sequenced and published in 2005. It has a high proportion of guanine and cytosine (69%) and an abundance of insertion sequences, with IS900 being the first to be discovered in any species mycobacteria and now generally recognized as unique to MAP. This 1451 base-pair long segment of DNA occurs in 15-18 copies per MAP cell and was independently identified by two laboratories in 1989 (DM Collins et al. 1989, and EP Green et al., 1989).
The DNA of MAP is >99% identical with that of M. avium, hence the change in naming as described above. The K-10 strain of MAP was fully sequenced and published in 2005. It has a high proportion of guanine and cytosine (69%) and an abundance of insertion sequences, with IS900 being the first to be discovered in any species mycobacteria and now generally recognized as unique to MAP. This 1451 base-pair long segment of DNA occurs in 15-18 copies per MAP cell and was independently identified by two laboratories in 1989 (DM Collins et al. 1989, and EP Green et al., 1989).
Genetic probes used for detection of MAP in clinical specimens or identification of MAP in cultures are often based on detection of IS900. Other commonly used genetic markers unique to MAP are Mav2 and HspX. Several of these genetic targets are used in commercial PCR kits for diagnosis of paratuberculosis. Another insertion sequence, named IS901, defines mycobacterial isolates as M. avium subsp. avium (MAA). How these insertion elements affect the biology and pathogenic capacity of MAP or MAA is not understood. Evidence suggests, however, that they play a major role. Readers are directed to the article of Hsu et al, 2011 for the latest detailed information on the MAP genome and the 2008 article by Turenne et al. regarding the evolution of both MAP and MAA from their MAH ancestor.