Antonio Foddai and Irene Grant from the Institute for Global Food Security, School of Biological Sciences, Queen’s University Belfast, Belfast, Northern Ireland have created a novel assay to quantify viable MAP in milk (patent pending). Their report, titled “A novel one-day phage-based test for rapid detection and enumeration of viable Mycobacterium avium subsp. paratuberculosis in cows’ milk” appeared September 24 in the journal Applied Microbiology and Biotechnology [Open Access].
In this assay phages (viruses that infect bacteria) are used to both concentrate MAP and to then infect and lyse MAP, allowing for final detection and quantification by qPCR. The total assay time is roughly 7 hours, faster than other phage-based assay systems. The key to the assay is the binding orientation of the phage. The head is bound to the paramagnetic bead allowing the tail to bind to MAP. After binding, the phage then lyses the MAP organism making its DNA available for detection by qPCR.
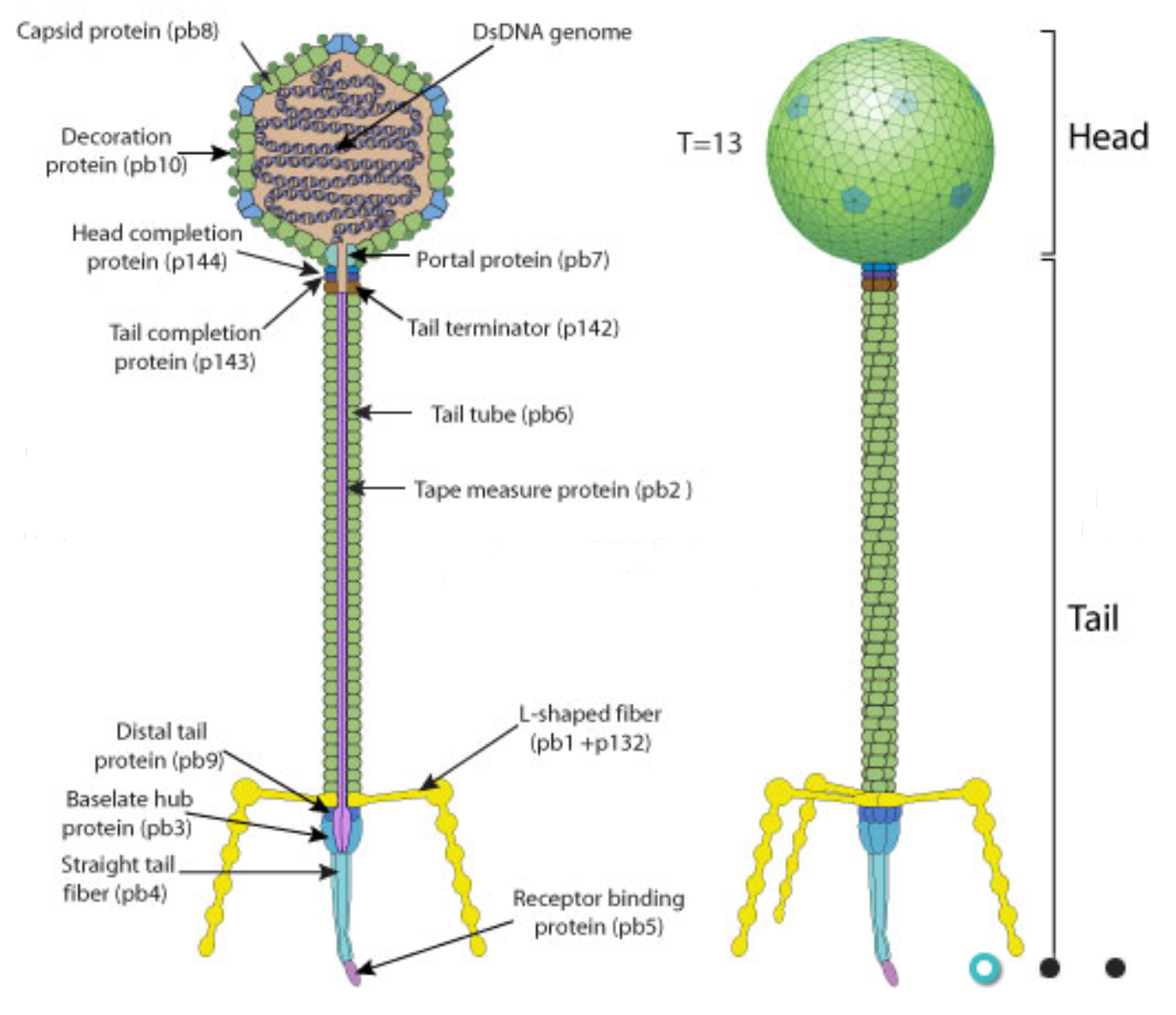
Diagram of a similar phage from ViralZone.
Abstract
Bacteriophage-based methods for the rapid detection of viable Mycobacterium avium subsp. paratuberculosis (MAP) in veterinary specimens are a recent addition to the Johne’s disease diagnostic toolbox. Here, we report the use of D29 mycobacteriophage-coated tosylactivated paramagnetic beads to capture and concentrate MAP cells from samples (termed phagomagnetic separation, PhMS) and then naturally lyse viable MAP cells (from the inside out) to provide DNA for IS900 qPCR purposes. Transmission electron microscopy confirmed that D29 phages had bound to beads in the correct orientation and that the phage-coated beads captured MAP cells from a suspension. During test optimization, conventional IS900 PCR results were used to subjectively assess the effect of different phage:bead coating ratios, differing amounts of coated beads during PhMS, optimal incubation time post-PhMS to obtain maximal MAP DNA, and the potential benefit of a brief heat shock (55 °C/1 min) prior to IS900 TaqMan qPCR. The limit of detection 50% (LOD50%) of the optimised PhMS-qPCR assay was 10.00MAP cells/50 ml milk (95% CI 1.20–82.83). Finally, in order to demonstrate the new assay’s ability to detect viable MAP in naturally contaminated milk, bulk tank milk samples from 100 dairy farms were tested. Forty-nine (49%) of these tested PhMS-qPCRpositive, with viable MAP numbers detected ranging from 3–126 MAP/50 ml. The novel PhMS-qPCR assay is a sensitive, specific and easy-to-apply phage-based assay for viable MAP, with potential application for milk surveillance or diagnosis of Johne’s disease.
Comment: No other assay system can claim the ability to quantify such low numbers of viable MAP in milk in such a short time. I predict that this assay will reveal that there is far more MAP in raw milk than previously recognized. Hopefully this novel technology also can be applied to other types of clinical samples, e.g. blood samples from Crohn’s disease patients. For more about D29 phages see this Wikipedia page.