Research Article
Elvira Ramovic and 9 colleagues reported a pilot study to assess the rate of MAP infection in Irish dairy and beef cattle herds, and much more. The abstract is below and the comments that follow discuss the dairy survey prevalence data specifically. The article is in the Irish Veterinary Journal and is Open Access.
Abstract
Background: Dairy and beef cattle can be reservoirs of many pathogens, including Salmonella and Mycobacterium avium subsp. paratuberculosis (MAP), the causative agent of Johne’s disease (JD). Farm environments may provide potential entry points for the transmission of infectious agents into the food chain. Antibiotics are used to treat a wide variety of infections on farms, and administration of antimicrobial agents to cattle is considered to be a driving factor for antimicrobial resistance (AMR). Control of JD and AMR are priority for animal health initiatives in Ireland. A national JD pilot programme was introduced by Animal Health Ireland in 2014, while the national action plan launched by Department of Health and Department of Agriculture, Food and Marine introduced in 2017 aims to improve the surveillance of AMR. The current investigation was undertaken as a pilot study to determine the proportion of herds positive for MAP, Salmonella species (Salmonella spp), commensal Escherichia coli (E. coli), Extended-spectrum beta-lactamase (ESBL) AmpC β-lactamase and carbapenemase-producing E. coli from 157 environmental faecal samples in Irish farms.
Results: MAP was detected in 10.2% of samples collected; on culture in 4 (4.9%) of the dairy herds and from 1 (1.3%) of the beef/suckler herds, and by PCR in 10 (12.3%) and 6 (7.9%) of these herds respectively. All culture positive herds were also positive by PCR. An additional 11 herds were positive by PCR only. Salmonella was not detected, while commensal E. coli were isolated from 70.7% of the samples (111/157) with 101 of these isolates shown to be fully susceptible to all antimicrobials tested. Of the 27 presumptive ESBL AmpC β-lactamase producing E. coli detected, one isolate was resistant to ten antimicrobials, nine isolates were resistant to nine antimicrobials, and four isolates were resistant to eight antimicrobials. Carbapenemase-producing E. coli were not isolated.
Conclusions: The results highlight the importance of monitoring farm environments for Johne’s disease. This disease is a growing concern for dairy and beef producers in Ireland, and sampling the farm environment may offer a useful means to rapidly screen for the presence of MAP. Non-pathogenic common enteric commensal and multiple-drug-resistant E. coli may contribute to AMR acting as a reservoir and transferring resistance to other species/pathogens in the environment.
Comment: As they say, “the devil is in the details”.
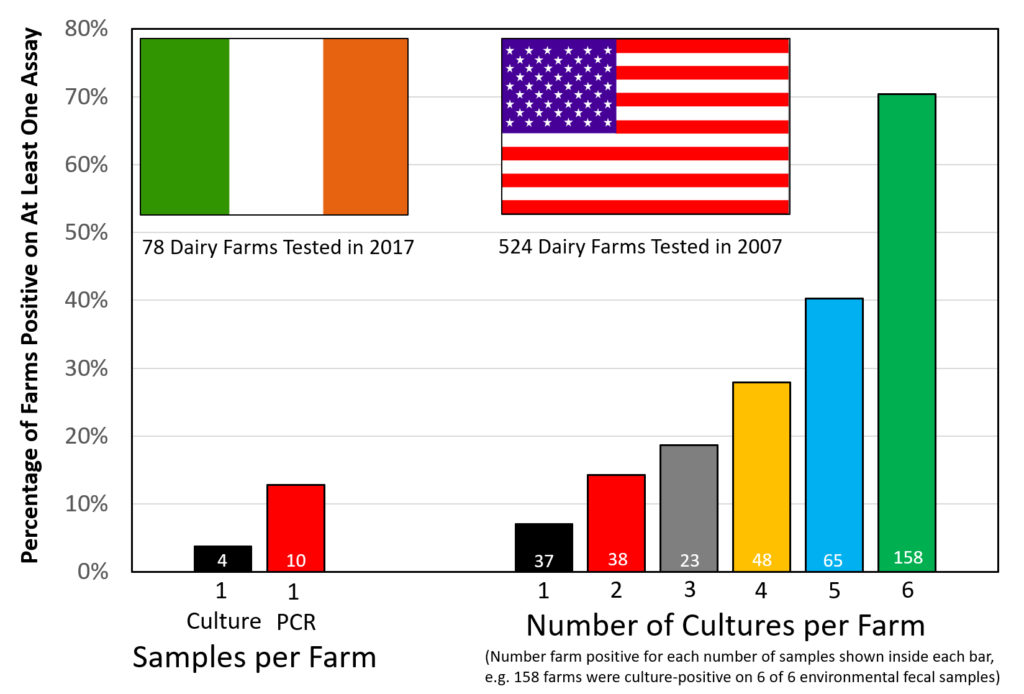
The above graphic contrasts the two similar surveys for MAP on dairy farms, both done using environmental fecal samples.
The Irish survey used culture methods on the first sample finding 4 farms positive of the 78 sampled (5.1%). The second sample was tested using IS900 PCR finding 10 of 78 (12.8%) positive. All 4 culture-positive farms were also PCR-positive. Were more farms detected because PCR is more sensitive, or because a second sample increases the chances of detecting an infected farm?
The US survey only used culture methods to judge the farm’s MAP infection status but collected 6 samples per farm. The US data indicates that: the more you look the more you find. Over 70% of US dairy farms had a least one culture-positive sample. From this Lombard et al. estimated the true herd-level MAP infection ate in US dairy herds to be 91.1%.
Clearly there are more MAP-infected farms in the U.S. than in Ireland, however, it is tricky to estimate just how many would have been detected if only 2 samples were tested, i.e. which 2 of the 6 tested samples would you count? I will leave that for the modelling experts. For the purposes of this news item it is not that important.
However, since this was a pilot study, it seems that a larger study is warranted that: 1) randomly selects farms, 2) uses PCR as the probably more sensitive and definitely more cost-effective diagnostic test, 3) measures the analytical sensitivity of the test based on spiked samples to better understand the accuracy of the measurement tool, and 4) collects more than 2 environmental fecal samples per farm.
For detection of MAP-infected dairy farms with a low infection prevalence, e.g. 0 to 12%, as in the Irish situation, Lombard et al. estimated the herd-level sensitivity of a single environmental fecal culture at 33.3% and two environmental fecal cultures at 44.4% (see Table 1 in Lombard, 2013). If Lombard et al. are correct, the Irish survey may have under-estimated the true MAP infection status of its dairy farms.
BUT to be fair – it was only a pilot study! (and nobody asked my opinion). 🙂