Antonio Foddai and colleagues have developed a novel assay for detection of viable MAP in milk. The assay uses bacteriophages (viruses that attack bacterial cells) attached to magnetic beads to capture MAP in milk samples and then detect the presence of MAP by PCR. While these phages bind to both living and MAP cells, they only then infect and lyse live MAP. Thus, this assay only detects live MAP, just as do conventional culture methods. This new assay has higher diagnostic sensitivity and specificity than the most commonly used assay on milk today, the milk ELISA.
The article, published in the Journal of Dairy Science, is not Open Access, but is available for the next 40 days.
ABSTRACT
Bulk tank milk samples from 392 Northern Ireland dairy farms and individual milk from animals (n = 293) on 4 of these farms were tested by a novel phagomagnetic separation (PhMS)-quantitative (q)PCR assay able to detect and quantify viable Mycobacterium avium ssp. paratuberculosis (MAP), to demonstrate its potential utility as a milk surveillance tool. Viable MAP were detected in 26.5% of the bulk tank milks, with MAP contamination levels ranging from 1 to 8,432 MAP/50 mL of milk; less than 2% of farms had MAP contamination levels >100 MAP/50 mL in their bulk tank milk. Follow-up PhMS-qPCR testing of milk from individual animals on 4 farms that had the highest numbers of MAP in their bulk tank milks indicated that 17 to 24% of animals in each herd were shedding viable MAP in their milk. Mean MAP numbers detected ranged between 6.7 and 42.1 MAP/50 mL of milk. No significant correlation was observed between the detection of viable MAP in bulk or individual milks by PhMS-qPCR and parallel milk ELISA results, or between PhMS-qPCR results and any other milk recording results (somatic cell count, total bacterial count, % butterfat, or % protein). Viable MAP was detected by IS900 qPCR in 52 (85.2%) Pozzato broth cultures of 61 PhMS-qPCR-positive individual milks after 12 wk of incubation, suggesting few PhMS-qPCR results were false positives. The mean sensitivities of the PhMS-qPCR assay and milk ELISA applied to individual milks were estimated by Bayesian latent class analysis to be 0.7096 and 0.2665, respectively, and mean specificities were similar (0.9626 and 0.9509). Our findings clearly demonstrate that the novel PhMS-qPCR assay could be a useful milk surveillance tool for dairy processors, or a milk monitoring tool for Johne’s disease control or milk quality assurance programs.
COMMENT
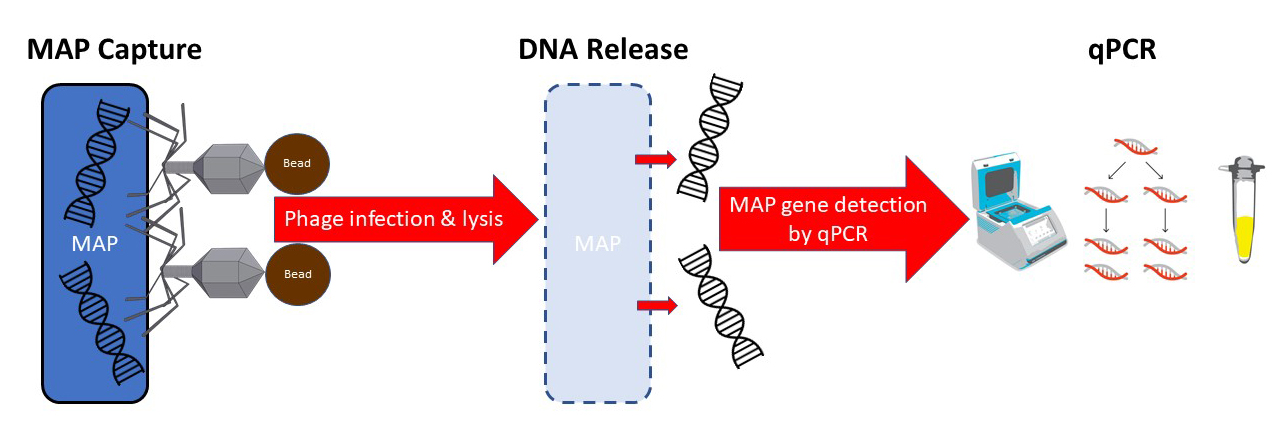
Phage-based assays for MAP have been described previously. This is the first time, however, that the phage has been linked to a magnetic bead allowing retrieval and concentration of MAP using magnets. In addition to binding to MAP, the phage then infects and lyses the live MAP bacterial cell, releasing its DNA allowing detection by qPCR technology. My schematic of the process appears below.
The head of the phage is linked to a magnetic bead. The tail of the phage binds to live mycobacteria. A magnet then pulls all of the beads+phage+MAP out of the sample.
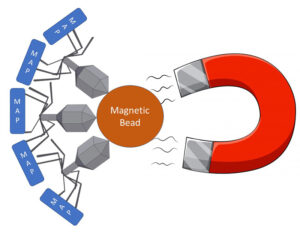
After collecting the beads on a magnet, the phage then infects and lysis the live mycobacterial cells releasing their DNA. A qPCR specific for MAP then detects the pathogen. The phage-bead-magnet combination makes the assay very sensitive and the qPCR part of the assay makes it very MAP-specific.
Phage amplification assays for detection of mycobacterial infections were recently reviewed in the journal Microorganisms. This excellent 28 page Open Access article with 97 references provides a deeper understanding of phage-based mycobacterial detection technology but does not address this new twist: the use of phages to both capture and lyse the mycobacteria.
Actiphage is a commercial test, based on phage technology, for diagnosis of bovine tuberculosis and Johne’s disease. Dr. Cath Rees (University of Nottingham) will be presenting a talk “Use of Actiphage to improve our understanding of mycobacterial diseases” on the 23rd March at 2pm as part of a virtual virology conference. Registration for the event is free.
The Wisconsin Connection
Fun factoid: Dr. Tom Brock, University of Wisconsin-Madison Professor, discovered a bacterium he named Thermus aquaticus, which grows at temperatures over 70 degrees C (158 degrees Fahrenheit) . Parts of its metabolic machinery, including the enzyme Taq polymerase, needed to assemble strands of DNA, tolerates boiling. In 1989, Science named Taq polymerase its first “molecule of the year,” because it enabled polymerase chain reaction (PCR), an essential tool for molecular biologists. Today’s #1 test for COVID also depends on PCR.
The world owes a big “thank you” to Dr. Tom Brock.