Research Poster
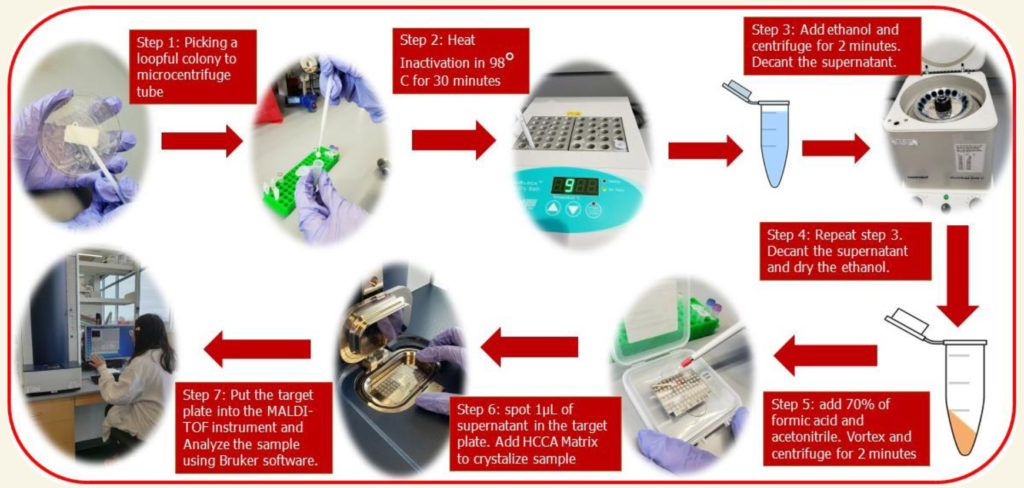
Undergraduate student, Claudia Antonika, at the University of Nebraska-Lincoln, under the direction of Dr. John D. Loy, in the School of Veterinary Medicine and Biomedical Sciences, evaluated the ability of the Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-ToF) to distinguish Mycobacterium avium subspecies paratuberculosis (MAP) from other members of the Mycobacterium Avium Complex (MAC), such as Mycobacterium avium subspecies hominissuis (MAH) and Mycobacterium avium subspecies avium (MAA) using the Bruker Biotyper MAlDI-ToF instrument (pictured below) and software. This research was presented as a poster at the University of Nebraska and is available here.

Results: MALDI spectral profiles evaluated by the Bruker Daltonics (BDAL) database only classified 20% (9/45) of M. avium subspecies correctly. In contrast, when the spectral patterns were matched against a new main consensus library (MSP database), 84% (38/45) of M. avium were accurately identified at the subspecies level, including 93% (26/28) MAP.
Conclusions: MALDI-ToF can improve the ability of labs to rapidly identify multiple M. avium subspecies. The new library database created in this project significantly enhances the specificity of the instrument as it increases the matching score values. Kappa analysis showed good strength agreement. Over 84% of the isolates could be classified correctly using our new MSP database, whereas current database BDAL only could identify 20% of MAC isolates. Although there were seven isolates that failed to have higher score values when using our MSP database, their top match values were still greater as compared to BDAL database score. Therefore, we consider that our new MSP database is a large improvement over the previous database.
Credits: This Poster is brought to you for free and open access by the UCARE: Undergraduate Creative Activities & Research Experiences at DigitalCommons@University of Nebraska – Lincoln. It has been accepted for inclusion in UCARE Research Products by an authorized administrator of DigitalCommons@University of Nebraska – Lincoln.
Comment: This is the largest study of MAP identification using MALDI-ToF, the current standard for bacterial identification in clinical microbiology laboratories. It provides an improved database with which to compare new M. avium isolates greatly improving identification accuracy. Moreover, it provides a new method for MAP identification that is independent of the genetics of MAP isolates, i.e. does not use genetic targets such as IS900 or HspX, to identify this M. avium subspecies. The only other main genetic-independent methods are: 1) exceptionally slow growth and 2) in vitro dependence on mycobactin for growth.