HAPPY BIRTHDAY DR. JOHNE
2022-12-10 16:25:58 One-hundred eighty-three years ago, on December 10, 1839, Heinrich Albert Johne was born in Dresden, Germany. It seems fitting that Johnes.org celebrate, on this date, his lasting contribution to veterinary medicine.
One-hundred eighty-three years ago, on December 10, 1839, Heinrich Albert Johne was born in Dresden, Germany. It seems fitting that Johnes.org celebrate, on this date, his lasting contribution to veterinary medicine.
Below you will find the story of his discovery, a brief biography, a little about the pathogen name, and the story of how I found this photo.
How it all started.
 Dr. F. Harmes, a veterinarian in the Oldenburg region of Germany in 1895, had a client with a Guernsey cow that was doing poorly. Dr. Harmes’ preliminary diagnosis was intestinal tuberculosis (TB). TB in cattle was quite common in Germany then. But when he did the tuberculin skin test to confirm his diagnosis, the cow tested negative. So, the reason for the cow’s condition remained a mystery.
Dr. F. Harmes, a veterinarian in the Oldenburg region of Germany in 1895, had a client with a Guernsey cow that was doing poorly. Dr. Harmes’ preliminary diagnosis was intestinal tuberculosis (TB). TB in cattle was quite common in Germany then. But when he did the tuberculin skin test to confirm his diagnosis, the cow tested negative. So, the reason for the cow’s condition remained a mystery.
A few months later, the cow died. Curious as to what killed the cow, Dr. Harmes sent intestines and other tissues to the Pathology Unit at the veterinary school in Dresden. There the tissues were examined by Dr. Heinrich A. Johne, Professor of Pathology, and Dr. Langdon Frothingham, a visiting scientist from the Pathology Unit in Boston, Massachusetts.
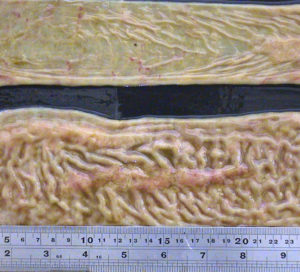 They observed that the small intestine was quite a bit thicker than expected and that lymph nodes near this thick intestine were enlarged. The photo at the right shows a normal intestine at the top and the intestine thickened due to Johne’s disease at the bottom. Lymphoid tissue, called Peyer’s Patches, are also quite prominent (the raised and slightly red tissue running long-ways down the center of the thickened intestine). Interestingly, Dalziel in 1913 saw the same kind of pathology when he removed a section of intestine from a person with Crohn’s disease remarking in his report that it resembled the cattle problem Dr. Johne had described.
They observed that the small intestine was quite a bit thicker than expected and that lymph nodes near this thick intestine were enlarged. The photo at the right shows a normal intestine at the top and the intestine thickened due to Johne’s disease at the bottom. Lymphoid tissue, called Peyer’s Patches, are also quite prominent (the raised and slightly red tissue running long-ways down the center of the thickened intestine). Interestingly, Dalziel in 1913 saw the same kind of pathology when he removed a section of intestine from a person with Crohn’s disease remarking in his report that it resembled the cattle problem Dr. Johne had described.
Using what at the time were newly developed histopathology techniques, parts of the intestine were “fixed” (pickled in formaldehyde), sliced into very thin sections, placed on a microscope slide, and stained with special dyes – known as an acid-fast stain - designed to help visualize bacteria of the type causing TB.
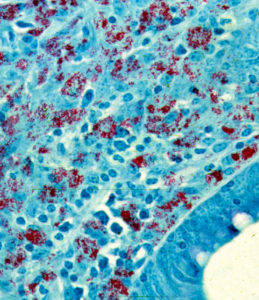
Under the microscope, Drs. Johne and Frothingham saw that the intestinal wall was filled with inflammatory cells of the kind to be expected in TB (macrophages and lymphocytes – the blue-colored stuff in the photo). In addition, they saw abundant red-staining bacteria (which microbiologists call acid-fast bacteria) throughout the inflamed tissues. Basically, it looked just like intestinal TB. But, when a sample of the fresh infected tissue containing the red-staining bacteria was injected into guinea pigs, it didn’t cause TB. This took place shortly after Louis Pasteur had devised the “germ theory” of disease and before techniques for growing bacteria in the laboratory were widely available. Inoculating animals, therefore, was a routine way of detecting infectious microbes such as those that cause TB, and guinea pigs are quite susceptible to tuberculosis. So, the diagnosis on this cow remained a mystery.
Drs. Johne and Frothingham concluded that the disease seen in the very sick Guernsey cow was caused by a bacterium other than the one normally causing TB in cattle, namely Mycobacterium bovis. They speculated that perhaps the pathology was due to a related bacterial pathogen such as the one causing TB in birds, aptly named Mycobacterium avium. Considering their subject’s gross pathology, microscopic pathology (histopathology) and animal inoculation findings, they proposed the name "pseudotuberculous enteritis" for the disease; a designation meaning inflammation of the intestine resembling intestinal TB but not actually the same as intestinal TB – somehow different. Soon after publication of their report, veterinarians began reporting outbreaks of this curious intestinal malady among dairy cows in Denmark, The Netherlands and elsewhere in continental Europe.
More on Dr. Johne.
H.A. Johne was the son of a veterinarian. Twenty years later, he became a veterinarian himself and held a practice for the next seven years. From 1866-1876, he acted as district veterinary inspector. He was then appointed to a lectureship at the veterinary school in Dresden. For a teacher of veterinary medicine, he lectured in an unusually wide range of subjects: embryology, histology, obstetrics, exterior, physical diagnostics. In 1879, he was appointed professor of pathological anatomy and of general pathology. Later he also lectured on parasitology and methodical zoology, and he also started classes in such a new branch of research as bacteriology.
As a scientist, he concerned himself with tuberculosis, anthrax, rabies, glanders, actinomycosis, bothryomycosis among others. As a writer he left a wide literary production. His books were printed in a dozen editions. For many years he also edited “Zeitschrift tor Tiermedizin”, and acted as co-editor of “Rundschau auf dem Gebiet der Fleischbeschau”. In 1887, he visited Denmark, where he was nominated honorary member of the Danish Association of Veterinarians and decorated with the Order of Knight of the Dannebrog.
He was often guest of Professor B. Bang and his family, the flat of Bangs' is today the Veterinary History Museum, established in 1973 at the Royal Veterinary and Agricultural University in Copenhagen. His motto was: Duty Above All. With the distinguished array of titles: Geheim-Medizinalrat, Professor, Dr. med., Dr. med.vet.h.c. and Dr. phil., Heinrich Albert Johne retired in 1904, respected and honored by his many students and by foreign veterinary schools and societies. He died in 1910.
MAP
In 1912, in one of those curious discoveries by serendipity, Twort and Ingram discovered how to grow the cause of Johne’s disease in the laboratory and named this bacterial pathogen Mycobacterium enteritidis chronicae pseudotuberculosae bovis johne. Time and technology led to name changes and the cause of Johne’s disease is today known as Mycobacterium avium subspecies paratuberculosis or simply MAP. Johne’s disease, also called paratuberculosis, is now a disease of major global importance.
Dr. Johne photo credit
I found the photo of Dr. Johne was hanging in halls of the State Veterinary Serum Institute when I was on sabbatical working with Dr. J.B. Jørgensen at the State Veterinary Serum Laboratory, Copenhagen, Denmark. Together we were comparing new methods for culturing MAP from clinical samples. On my departure, Dr. Jørgensen gifted me a copy of this photo which hangs in my office and also appears on the Wikipedia page about Dr. Johne.
PS
For more historical events and people important to understanding of Johne’s disease visit our history timeline.
CROHN'S DISEASE
2022-08-15 14:48:10MAP has long been associated with Crohn’s disease and many patients have benefited from anti-MAP antibiotic therapy. You can read one such story where a veterinarian was fully cured of Crohn’s disease by anti-MAP antibiotic therapy here.
 Dr. Marcel Behr, Professor of Medicine at McGill University and Associate Director of the Infectious Diseases and Immunity in Global Health Program at the Research Institute of the McGill University Health Centre conducted an interview on research connecting Crohn’s disease to MAP. To me, it is one of the most thoughtful, objective assessments of the state of research on the MAP=CD hypothesis I have ever witnessed. The interview covers every aspect of the research on this association in a lucid, straight forward discussion of this controversial topic.
Dr. Marcel Behr, Professor of Medicine at McGill University and Associate Director of the Infectious Diseases and Immunity in Global Health Program at the Research Institute of the McGill University Health Centre conducted an interview on research connecting Crohn’s disease to MAP. To me, it is one of the most thoughtful, objective assessments of the state of research on the MAP=CD hypothesis I have ever witnessed. The interview covers every aspect of the research on this association in a lucid, straight forward discussion of this controversial topic.
The interview is on You Tube. I appeared online August 13, 2022. It is 57 minutes long and, due to its technical nature, is probably best appreciated by physicians, veterinarians, Crohn’s disease patients, and people who follow this line of research.
Those wanting more background on this subject should visit the page on this site titled Zoonotic Potential where a synopsis of the subject is presented together with a long list of the primary research papers related to the subject and direct links to the full publications.
MAP VS HUMANS
2022-06-03 15:06:27Dr. C.T. Dow and Ms Briana L. Alverez from the University of Wisconsin-Madison have published a review article in EcoHealth titled: Mycobacterium paratuberculosis zoonosis is a One Health emergency. The article is Open Access (free to everyone), well-written, succinct (7 pages plus references) and well-researched with 113 literature citations.
ABSTRACT
A singular pathogen has been killing animals, contaminating food and causing an array of human diseases. Mycobacterium avium subspecies paratuberculosis (MAP) is the cause of a fatal enteric infectious disease called Johne’s (Yo’-nees), a disorder mostly studied in ruminant animals. MAP is globally impacting animal health and imparting significant economic burden to animal agriculture. Confounding the management of Johne’s disease is that animals are typically infected as calves and while commonly not manifesting clinical disease for years, they shed MAP in their milk and feces in the interval. This has resulted in a ‘‘don’t test, don’t tell’’ scenario for the industry resulting in greater prevalence of Johne’s disease; furthermore, because MAP survives pasteurization, the contaminated food supply provides a source of exposure to humans. Indeed, greater than 90% of dairy herds in the US have MAP-infected animals within the herd. The same bacterium, MAP, is the putative cause of Crohn’s disease in humans. Countries historically isolated from importing/exporting ruminant animals and free of Johne’s disease subsequently acquired the disease as a consequence of opening trade with what proved to be infected animals. Crohn’s disease in those populations became a lagging indicator of MAP infection. Moreover, MAP is associated with an increasingly long list of human diseases.
Despite MAP scientists entreating regulatory agencies to designate MAP a ‘‘zoonotic agent,’’ it has not been forthcoming. One Health is a global endeavor applying an integrative health initiative that includes the environment, animals and humans; One Health asserts that stressors affecting one affects all three. Recognizing the impact MAP has on animal and human health as well as on the environment, it is time for One Health, as well as other global regulatory agencies, to recognize that MAP is causing an insidious slow-motion tsunami of zoonosis and implement public health mitigation.
COMMENT
The discussion section of this publication summarizes the situation best and so it is quoted here:
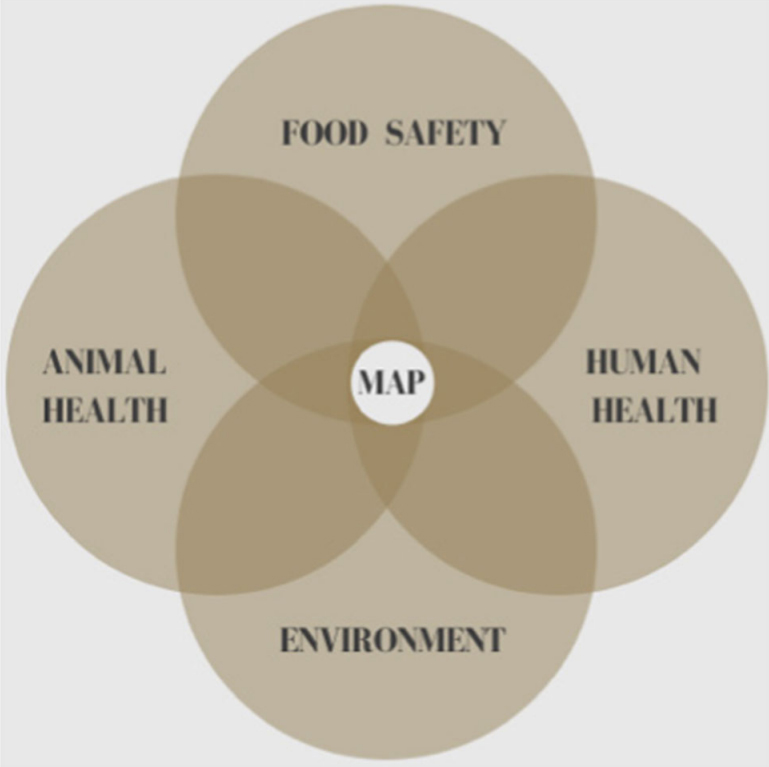
The incidence of T1D in children is increasing worldwide (Hummel et al. 2012) as is the incidence of Crohn’s disease (Torres et al. 2017). Both the principle of parsimony and Koch’s postulates support inculpation of MAP as a cause of Crohn’s disease. Regardless of the relative strength one might assign to the MAP/Crohn’s association, this article enumerates other MAP-associated diseases and the increasing medical literature supporting it (Ekundayo and Okoh 2020). The combined weight of these disease associations should incite a call to action by regulatory agencies to invoke the precautionary principle with regard to consumption of MAP-contaminated food in at-risk individuals. In spite of public health implications, contamination of milk and dairy products with MAP is not currently restricted. We view the management of public health risk due to MAP as an increasingly important policy issue. With mounting global recognition of the impact MAP has upon the health of the environment, animals and humans, One Health is well positioned at that nexus (Fig. 1). One Health is in a unique position to elevate the discussion to mitigate this emerged yet neglected zoonotic pathogen: Mycobacterium avium subspecies paratuberculosis.
IRISH JD REVIEW
2022-05-26 15:07:56N. Field from the Animal and Bioscience Research Department, Teagasc, Moorepark Research Centre, Fermoy, Co. Cork, Ireland and colleagues have published an excellent Open Access article concerning Johne’s disease in the Irish Veterinary Journal. The article is succinct and comprehensive with 99 references cited. The pathogenesis of Johne’s disease is described briefly followed by an excellent summary comparing the accuracy of diagnostic tests for Johne’s disease both at the individual cow-level and herd-level using the latest research findings.

ABSTRACT
Johne’s disease is an infectious disease affecting cattle, other ruminants and non-ruminant wildlife worldwide, caused by Mycobacterium avium subspecies paratuberculosis (MAP). This review provides an up-to-date concise overview of the pathogenesis of MAP, the significance of Johne’s disease in cattle and the use of diagnostic testing at both animal and herd level in the context of seasonal pasture-based herds. While MAP can only replicate intracellularly, the bacterium is sufficiently robust to survive for months in the environment. Transmission of MAP is mostly via the faecal-oral route, however in-utero transmission in also possible. The bacteria evade the immune system by persisting in macrophages in the small intestine submucosa, with this latent stage of infection lasting, in most cases, for at least two years before bacterial shedding and clinical signs begin. The slowly progressive nature of MAP infection, poor performance of diagnostic tests and management systems that expose susceptible calves to infection make control of Johne’s disease challenging, particularly in seasonal calving herds. Testing of individual animals provides little assurance for farmers and vets due to the poor sensitivity and, in the case of ELISA, imperfect specificity of the available tests. Repeated herd-level testing is utilised by the IJCP to detect infected herds, identify high risk animals, and provide increasing confidence that test-negative herds are free of infection. The IJCP aims to control the spread of Johne’s disease in cattle in Ireland, in order to protect non-infected herds, limit the economic and animal health impact of the disease, improve calf health and reassure markets of Johne’s disease control in Ireland.
COMMENT
Ireland is the latest country to implement a national Johne’s disease control program and their scientists are concurrently generating research of practical importance to help drive the program. This publication is just one of many excellent products in recent years. Read more about the Irish program here.
DUBLIN MEETING ON MAP
2022-05-05 15:21:20The 15th International Colloquium on Paratuberculosis (15-ICP) will be held in Dublin Ireland, June 12-16, 2022.
To date, there are 182 registrants, and it is looking to be a very enjoyable conference. If you haven’t registered yet it’s not too late. The organizers are delighted with the number of abstracts submitted and have put together a strong scientific program. There are six plenary speakers, all experts from around the world, including Marcel Behr, Kumi De Silva, Frank Griffin, Marian Price Carter, Vivek Kapur and Herman Barkema.
There are six different sessions in the Scientific Program:
- Pathogenomics, Genotyping and MAP diversity (10 Oral Presentations)
- Control programs and Education (8 Oral Presentations)
- Diagnostics and detection (12 Oral Presentations)
- Host response and immunology (10 Oral Presentations)
- Epidemiology and Economics (10 Oral Presentations)
- Public Health and MAP in the environment (8 Oral Presentations)
A new feature at this colloquium is that before each poster session, five poster presenters will give a 2-minute pitch to encourage people to view their poster and discuss their research further. There are a total of 84 posters. The full program is available on the meeting website.
For details on travelling to Ireland please visit the Irish Government website.
There is also an interesting social program and you can anticipate many impromptu meetings, dinners and outings providing a chance meet leaders in MAP research personally. If you haven’t been before it is great opportunity to meet up with people working in the same field. There will be several social events targeted at graduate students, future MAP research leaders.
There is a Facebook page and a Twitter page. Details of places to visit when you come to Ireland will be included on these pages as well as Breaking News about the Colloquium.
COMMENT
This is the one and only international meeting focused on paratuberculosis (Johne’s disease) and the causative agent, MAP. It is always an exceptional meeting and a chance to meet and discuss this chronic, infectious, zoonotic pathogen of global importance with world experts.
FIRST JD CASES IN MEXICAN ZOO
2022-04-03 16:11:04A.L. Hernández-Reyes from the National Autonomous University of Mexico (UNAM), School of Veterinary Medicine and Zootechnics and colleagues reported on the first cases of Johne’s disease in zoo animals in Mexico. Their Open Access report appears in the journal Veterinary World.
ABSTRACT
Background and Aim: Paratuberculosis (PTB) is an infectious disease that induces chronic enteritis in ruminants. It is caused by Mycobacterium avium subsp. paratuberculosis (MAP). In this study, we evaluated the presence of MAP using bacteriological, molecular, and anatomopathological studies, based on the clinical suspicion of PTB in a zoo, in an area housing 10 scimitar-horned oryx (Oryx dammah), five giraffes (Giraffa camelopardalis), and three blue wildebeests (Connochaetes taurinus).
Materials and Methods: From November 2016 to June 2017, fecal samples were collected from individuals of the three species on four occasions, resulting in a total of 56 fecal samples. In addition, five small intestine samples were collected from the necropsies of three adult scimitar-horned oryx females and two oryx calves. MAP identification was performed through isolation in Herrold’s medium with egg yolk, mycobactin, and sodium pyruvate, Ziehl–Neelsen staining, IS900 polymerase chain reaction (IS900 PCR), and anatomopathological examination of intestine samples.
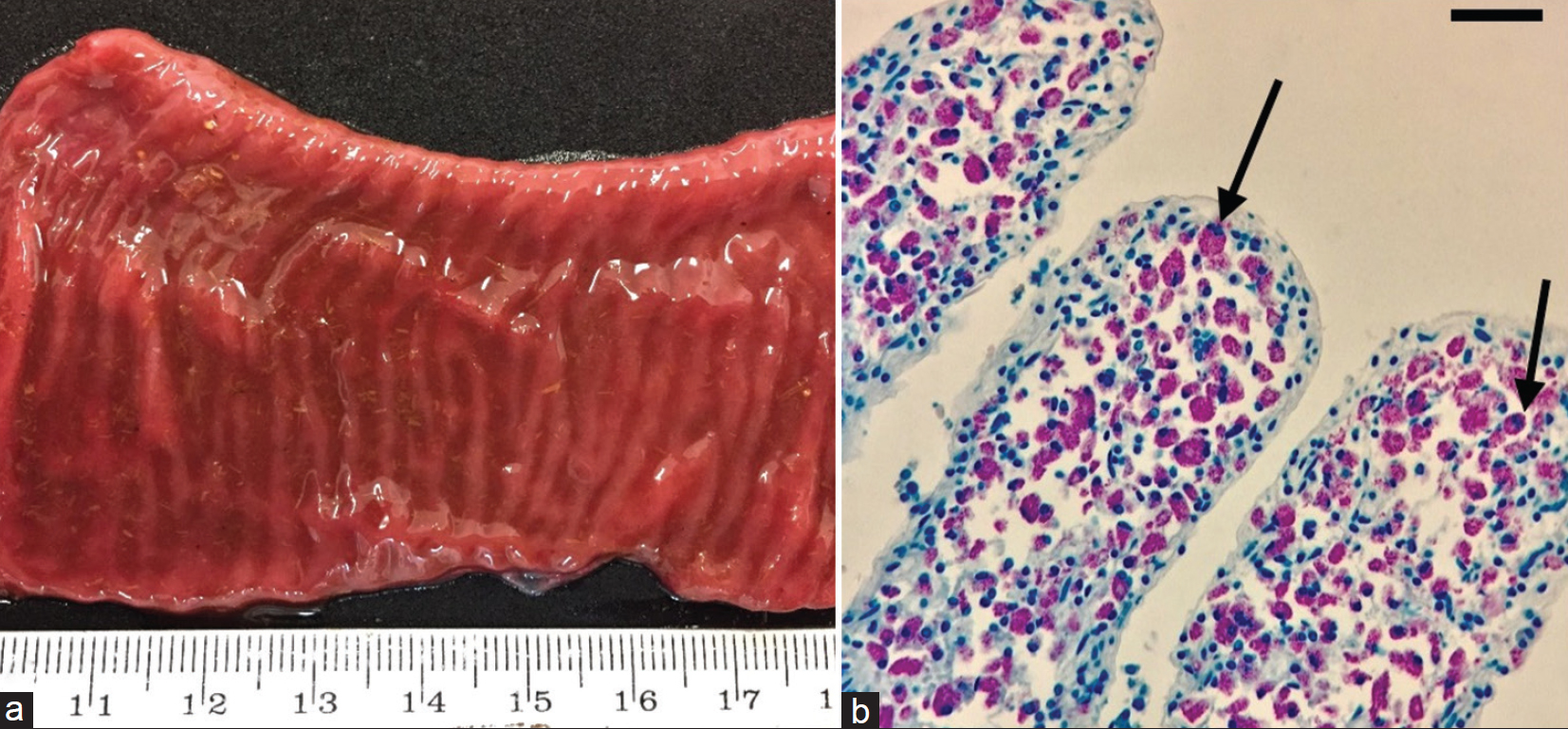
Results: Diffuse granulomatous enteritis with abundant acid-fast bacilli was found in two out of five intestine samples from adult scimitar-horned oryx females. MAP was isolated in 7/56 (12.5%) of the fecal samples from four scimitar-horned oryx, one giraffe, and two wildebeest samples. Two out of 5 (40%) samples obtained from scimitar-horned oryx tested positive. IS900 PCR yielded five positive samples (two fecal samples and three small intestine samples). MAP isolates were classified as Type C (Cattle) using type-specific PCR.

Conclusion: These results demonstrated the presence of MAP in the area evaluated and indicated the importance of both sampling live animals and conducting postmortem examinations. The use of bacteriological and histopathological diagnostic techniques demonstrated in this study will provide insight into the health status and prevalence of paratuberculosis in wild ruminants under human care.
COMMENT
Zoo veterinarians are keenly aware that MAP can infect and cause disease in many of the captive nondomestic ruminants they care for. For more about the efforts to control Johne’s disease in zoos and prevent exchange of MAP-infected animals among zoos I encourage you to go to this page on this website and follow links to several important publications on that page. Oryx photo credit: Wikipedia.
JD JOB IN IRELAND
2022-03-29 15:09:47 Animal Health Ireland (AHI) is seeking a Program Manager for their Johne’s disease (JD) control program.
Animal Health Ireland (AHI) is seeking a Program Manager for their Johne’s disease (JD) control program.
The following text is from their website:
Animal Health Ireland is a not-for-profit, public-private partnership, tasked with controlling livestock diseases not subject to international regulation, with the objective of improving the profitability and sustainability of Irish farmers and the agri-food sector. The advice provided by AHI is developed by a number of Technical Working Groups (TWGs) and the delivery of its major programmes, including one addressing Johne’s disease (JD), is overseen by cross-industry Implementation Groups (IGs).
The Position
The establishment of the Irish Johne’s Control Programme (IJCP) has been led by AHI in conjunction with industry stakeholders through the JD IG, guided by scientific input from the JD TWG. The IJCP is a long-term programme for the control of Johne’s disease and recognises the value of effective and on-going disease prevention and containment practices to control the infection. The IJCP provides pathways for test-negative and test-positive herds, while also improving calf health and biosecurity and providing market assurance.
A vacancy has arisen for a programme manager to oversee the continued development of the IJCP. Reporting directly to the CEO, the successful candidate will have responsibility for all aspects of the IJCP, building on the significant progress made to date, and will also have the opportunity to contribute to other AHI programmes and activities.
Follow this link for more details.
JD CONTROL SAVES CALVES
2022-02-20 15:55:13Control of Johne’s disease has direct benefits on calf health. This was well documented by K. Donat, E. Einax and A. Klassen in the state of Thuringian in Germany. Their recent publication appears in the journal Animals [Open Access].
The basis of this is that Johne’s disease is only one of several infectious diseases which are associated with lack of hygiene in calf rearing systems. Poor hygiene favors fecal-oral pathogen transmission. Measures that improve hygiene to prevent infections with MAP therefore also have positive effects on the occurrence of other fecal-orally transmitted infections such as Rota- and Coronavirus, Cryptosporidium parvum or Escherichia coli. Often these infectious agents cause severe clinical signs and mortalities in calves. Improvements in calf health are one additional reason for farmers to participate in Johne’s disease control programs.
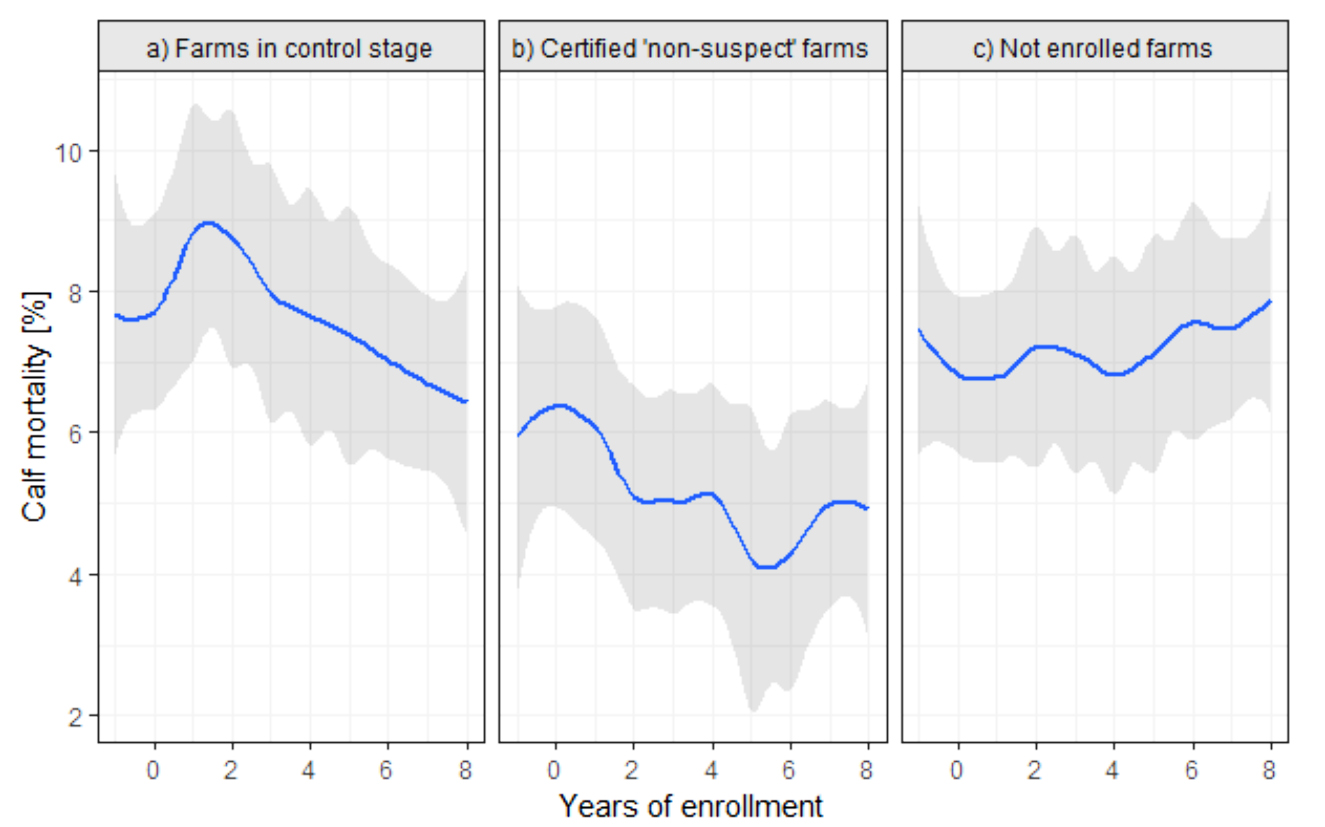
ABSTRACT
The Thuringian Johne’s Disease (JD) Control Program provides a voluntary approach to JD control in Thuringia, a federal state of Germany. The program has three objectives: reduce the level of infection when present; reduce the spread of JD to uninfected herds; and facilitate the certification and protection of herds that are non-suspect with respect to JD. The program offers pathways for the management of affected herds and for certification of herds with continuing negative tests. After the control stage (CS), a certification stage of at least 3 consecutive years with continuing negative results in the annual whole-herd test has to be passed until a herd can be certified as ‘non-suspect’ with respect to JD. This study focused on calf mortality in relation to JD herd status. In a longitudinal study, the association of annual calf mortality rate of a total of 93 dairy herds (13 ‘non-suspect’; 26 in control stage; 54 not enrolled) over 10 consecutive years with JD herd status was investigated using a generalized mixed linear model with repeated measures. Non-suspect herds had a lower calf mortality rate compared with other farms. We conclude that establishing JD control measures lowers the calf mortality rate.
COMMENT
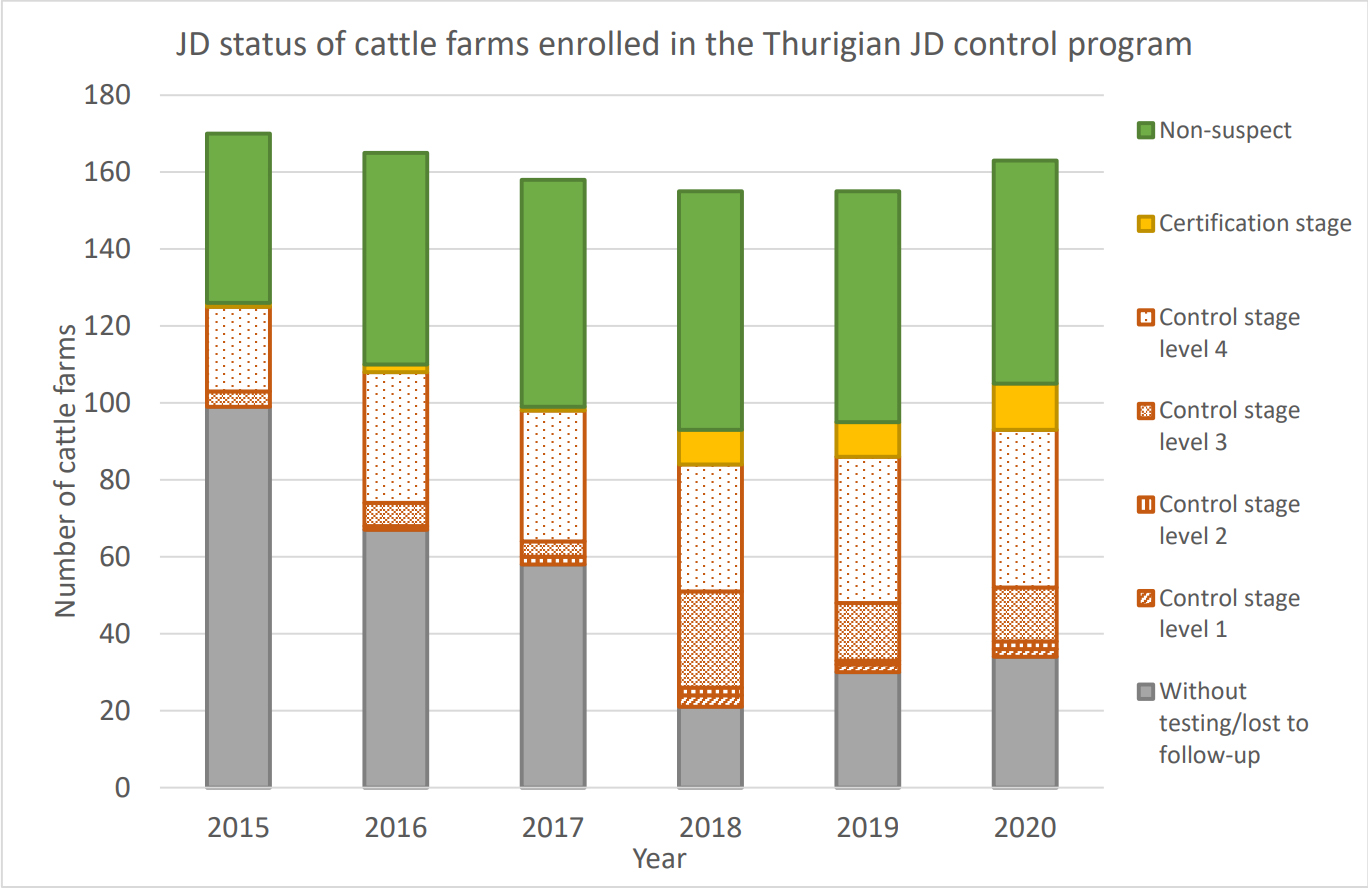
The Johne’s control program in Thuringian began in 2003 and was updated in 2008 and 2015. The graphic below shows the programs progress from 2015 to 2020. Germany is among the several countries with active Johne’s disease control programs and researchers are capitalizing on this to advance our understanding of Johne’s disease in dairy cattle.
Similar benefits to calf health by adoption of Johne’s disease control measures were shown in a Canadian study published in the Journal of Dairy Science in 2020. Calf health improvement is the first evidence that a JD control program is working. The benefits of higher milk production and lower culling rates come after three years or more.
MAP CAN TRIGGER MS
2022-02-13 21:41:23T.C. Ekundayo and colleagues have published a systematic review and meta-analysis on MAP as a trigger for multiple sclerosis (MS). The article (in press) appears in the journal Multiple Sclerosis and Related Disorders. The abstract appears toward the end of this news post. For non-experts to fully understand this study and its implications first requires some background information. Links are provided for those wanting more subject matter depth.
Multiple Sclerosis (MS) – extracted from Wikipedia
MS is an autoimmune disease (the body attacks itself) and is the most common demyelinating disease. Myelin in the material that provides insulation around nerve fibers and when it is lost (demyelination) the nerves cannot effectively transmit signals. The clinical signs of MS vary depending on which nerves are affected. Sensations such as tingling, or numbness and muscle weakness are common signs when peripheral nerves are affected. Vision and psychological problems also can occur when nerves in the brain are affected. Symptoms occur either as episodes of sudden worsening that last a few days to months (called relapses or flare-ups) followed by improvement (85% of cases) or as a gradual worsening over time without periods of recovery (10–15% of cases).
The cause of MS is unknown but believed to occur because of some combination of genetic and environmental factors such as infectious agents. MS is more common in people who live farther from the equator. Environmental factors may play a role during childhood. People who move to a different region of the world before the age of 15 acquire the new region's risk to MS. This is worth noting since there is an age-dependent susceptibility of animals, and quite likely humans, to MAP infection.
MS is not considered a hereditary disease; however, a number of genetic variations have been shown to increase the risk. Some genes linked to MS have also been implicated in other autoimmune diseases such as Type 1 Diabetes (another disease linked to MAP).
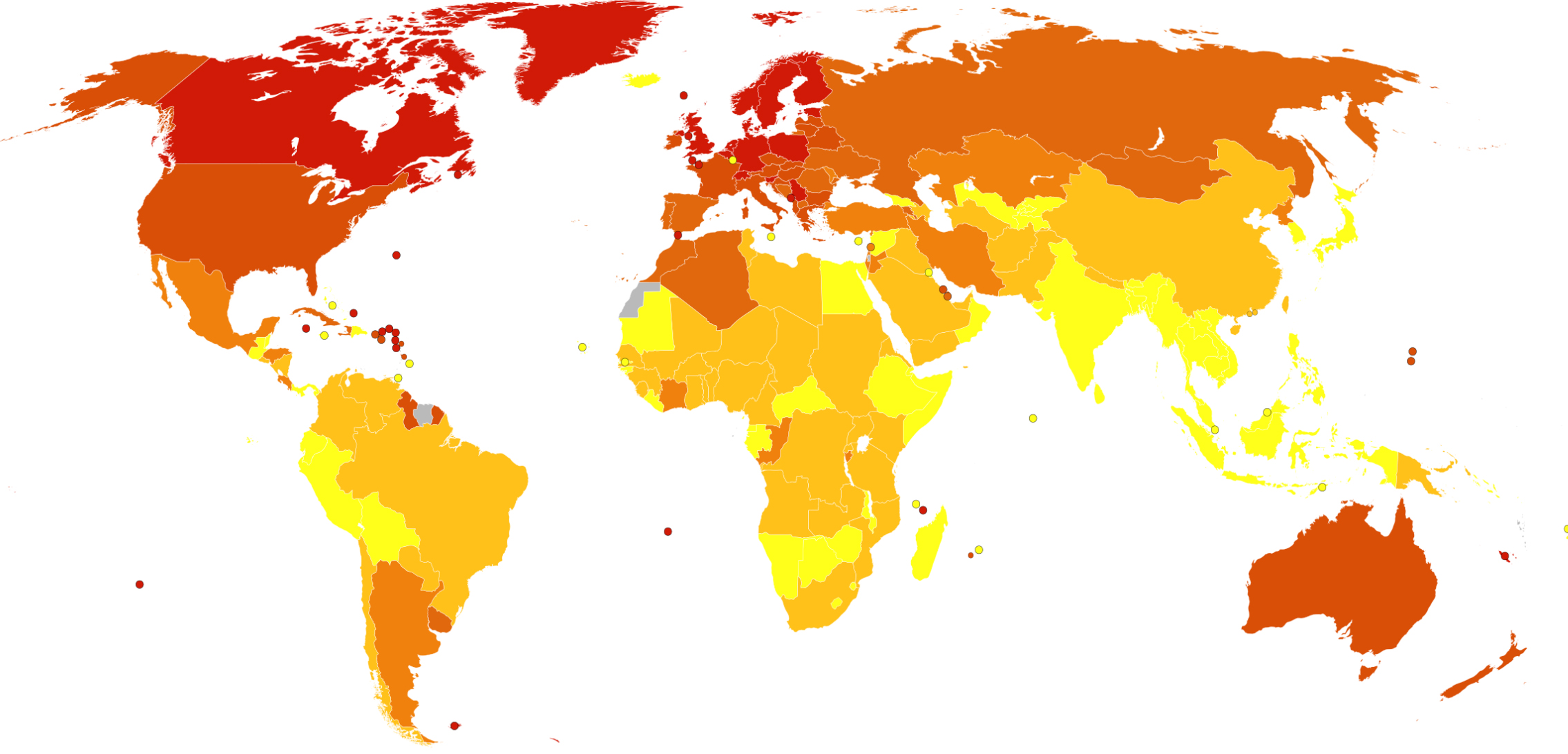
MS usually appears in adults in their late 20s or early 30s. As of 2010, the number of people with MS was 2–2.5 million (approximately 30 per 100,000) globally, with rates varying widely in different regions. The map below from Wikipedia shows death rates from MS per million people by country. The incidence of MS appears to be increasing.
Autoimmune diseases with bacterial triggers
The details of how and why microbes can trigger autoimmunity are complex and not fully agreed on. Molecular mimicry is one of the better understood mechanisms. In this situation, a microbe produces an antigen that shares structural similarities with antigens on specific host tissues. A chemical component of the pathogen mimics the structure of antigens on human tissues and the immune system fails to distinguish between the microbe and the host’s own tissues. When antibodies produced against this mimic microbial antigen bind to the host tissues, inflammation and disease can result. Rheumatic fever, which follows infection of people, most often children, with Group A Streptococcus bacteria (commonly known as “Strep throat”), is a well-known example. Rheumatic fever can be prevented by prompt treatment of Strep throat with appropriate antibiotics. Rheumatic fever is well-controlled in developed countries but remains a scourge elsewhere.
Food poisoning caused by Campylobacter jejuni (most often associated with under cooked poultry) is an interesting example because it also involves autoimmunity and the nervous system, as in MS. C. jejuni is one of the most common causes of food poisoning in Europe and in the United States. After a C. jenuni infection, a small proportion of people produce white blood cells (T-lymphocytes) and antibodies direct against parts of the C. jejuni cell wall that structurally resemble chemicals surrounding human nerves. When these antibodies mistakenly bind to the insulating material around nerve cells, inflammation occurs leading to muscle paralysis which is a condition called Guillain-Barré syndrome (GBS). The paralysis is typically reversible; nonetheless, about 20% of patients with GBS are left disabled, and around 5% die. Campylobacter infections can occur in all age groups. Studies show a peak incidence in children younger than 1 year and in people aged 15–29 years. Only about 1 in 2,000 people with a C. jejuni infection develop Guillain-Barré syndrome; essentially a combination of host genetics, level of immunity to C. jejuni infection from prior infections, C. jejuni biology, and bad luck. Because of the impact on human health, the poultry industry works hard to limit C. jejuni contamination of poultry products in the slaughterhouse (abattoir) and regulatory agencies, such as the USDA, monitor C. jejuni levels in retail poultry products.
MAP as a trigger for MS
Back to the topic of MAP and MS. T.C. Ekundayo and colleagues conducted a systematic review and meta-analysis on the hypothesis that MAP is a trigger for multiple sclerosis (MS). In lay terms, this means that they gathered all the available published scientific literature on the subject using specific scientific methodology and then used well-define statistical methods for judging the outcomes of those studies; essentially looking for scientific consensus. Such studies are daunting to read and understand for non-experts. However, the conclusions should be given considerable weight because they are the compilation of many studies done in different countries by diverse scientists. The authors concluded that there is a “strong association between MAP and MS, indicating that MAP is a significant environmental agent that may trigger MS.” The abstract of the publication is below and truly interested readers are encouraged to seek out the primary literature cited in the 42 references listed at the end of the publication.
ABSTRACT
Mycobacterium avium subsp. paratuberculosis (MAP) has been identified as one of the environmental infectious agents that causes multiple sclerosis (MS). The global prevalence of MS has been up surging over the years; however, efforts to divulge the role of MAP in MS have been limited. As a result, the present study aimed at assessing the odd ratios (ORs) associated MAP with the risk of MS. MAP-related MS data were obtained from 6 databases using the terms 'multiple sclerosis' or 'MS' and 'paratuberculosis' without regard for time or language restrictions following PRISMA standards. A total of 2,538 participants' data from 12 studies presenting anti-MAP antibodies and MAP DNA from 4 studies were fitted in random-effects (RE) and fixed-effects (FE) meta-analytic models. Furthermore, the between-study heterogeneity was measured using I2-values with a significant limit set at an I² > 75%. Analytical rigour and publication bias was determined using leave-one-out-analytics, Egger's tests, and p-curve analysis. In the FE and RE models, anti-MAP antibodies data significantly associated MS risk with MAP as 10.71 OR (95%-CI [7.78; 14.74], p-value < 0.0001) and 12.76 OR (95%-CI [8.13; 20.02], p-value < 0.0001) respectively, with an I2 value of 34.9% (95%-CI [0.0%; 67.2%]; p-value = 0.11). Similarly, the MAP DNA dataset in FE significantly present MS risk due to MAP as 5.53 OR (95%-CI [3.54; 8.66], p-value< 0.0001) while, RE showed 5.27 OR (95%-CI [3.22; 8.60], p = 0.0017), with an I2-value = 0.0% (95%- CI [0.0%; 84.7%]; p-value = 0.71). Eggers' test, on the other hand, found publication bias in anti-MAP antibodies data (intercept = 1.61, 95% CI: 0.45 – 2.77, t = 2.72, p = 0.021), but not in MAP DNA dataset (intercept = -5.57, 95% CI: -20.44 – 9.29, t = -0.74, p = 0.54). The robustness of the meta-analyses was demonstrated by all sensitivity analyses. In addition, there is no evidence of p-hacking observed (right-skewness test (PFull < 0.001, PHalf <0.001; statistical power ≥ 94% (95%-CI: 72.5%-99%)). In conclusion, the synthesis revealed a strong association between MAP and MS, indicating that MAP is a significant environmental agent that may trigger MS. Thus, early screening of MAP in MS cases may assist in the therapeutic approach to its management/treatment. Therefore, future studies should be tailored towards the role of MAP in the severity of MS phenotypes, as well as address global data gaps and low disease surveillance.
COMMENTS
The idea that MAP is a cause of multiple autoimmune diseases is plausible and backed by scientific data. Because MAP is in food (dairy and meat products primarily) and domestic water supplies, most humans are regularly exposed. For more on this see the page on MAP in food and water on this website.
As the MAP epidemic in food-producing animals has expanded over the past century, so too has the frequency of autoimmune disease in humans. There are global maps of where autoimmune diseases such as MS, Crohn’s disease, Type 1 Diabetes and others occur most commonly. These maps generally coincide with similar global maps showing the prevalence of MAP infections in animals (see the page on Zoonotic Potential). Whether this is a causal or coincidental relationship remains to be firmly established or accepted by the medical community. As an interesting added bit of information supporting a causal role of MAP, Dr. C.T. Dow recently reported that “Epidemiologic evidence points to BCG (a vaccine for tuberculosis) providing a “heterologous” protective effect on assorted autoimmune diseases; studies using BCG vaccination for T1D (Type I Diabetes) and MS have shown benefit in these diseases (Microorganisms, 2020).
Sadly, despite mounting evidence that MAP affects human health in diverse ways, the problem is largely being ignored. Public health agencies such as NIH and CDC and regulatory authorities such as the FDA and USDA have not labeled MAP a food safety, water safety, or public health problem despite finding live MAP in retail foods, detecting MAP in domestic water supplies, finding MAP in Crohn’s disease patients by culture-based methods and PCR, or as immune responses to MAP infection in Type 1 Diabetes and MS.
This creates a circular problem: no acknowledgment that MAP is a human health issue results in little or no research funding and thus no definitive answers as to the ongoing question of MAP and human health. Without funding, researchers can’t unravel this potentially huge and exceedingly complex problem. Agencies funding animal health research have limited funds and do not expend those precious funds on human health research. Agencies that fund human health research do not recognize MAP as a human pathogen making it difficult for the research community to get funding to conduct the necessary research. Moreover, the problem requires a team of scientists with MAP expertise working in close collaboration with specialists in the various human diseases of concern; a One Health approach. Only if such funding bodies openly recognized MAP as a potential human health problem will One Health research teams have success at securing the necessary funding to unravel the impacts of MAP on humans and find rational solutions for the future.
MAP IN GOATS & SHEEP
2021-12-26 21:34:01Sanaa M. Idris and colleagues published a review titled: Paratuberculosis: The Hidden Killer of Small Ruminants. The article appears in the journal Animals and is Open Access. This overview attempts to highlight the current research and gaps on this disease in small ruminants to draw more attention for further studies on diagnosis, prevention and control.
ABSTRACT
Paratuberculosis (PTB) is a contagious and chronic enteric disease of ruminants and many non-ruminants caused by Mycobacterium avium subsp. paratuberculosis (MAP), and is characterized by diarrhea and progressive emaciation with consequent serious economic losses due to death, early culling, and reduced productivity. In addition, indirect economic losses may arise from trade restrictions. Besides being a production limiting disease, PTB is a potential zoonosis; MAP has been isolated from Crohn’s disease patients and was associated with other human diseases, such as rheumatoid arthritis, Hashimoto’s thyroiditis, Type 1 diabetes, and multiple sclerosis. Paratuberculosis in sheep and goats may be globally distributed though information on the prevalence and economic impact in many developing countries seem to be scanty. Goats are more susceptible to infection than sheep and both species are likely to develop the clinical disease. Ingestion of feed and water contaminated with feces of MAP-positive animals is the common route of infection, which then spreads horizontally and vertically. In African countries, PTB has been described as a “neglected disease”, and in small ruminants, which support the livelihood of people in rural areas and poor communities, the disease was rarely reported. Prevention and control of small ruminants’ PTB is difficult because diagnostic assays demonstrate poor sensitivity early in the disease process, in addition to the difficulties in identifying subclinically infected animals. Further studies are needed to provide more insight on molecular epidemiology, transmission, and impact on other animals or humans, socio-economic aspects, prevention and control of small ruminant PTB.
COMMENT
Paratuberculosis in goats and sheep does not receive sufficient attention or research funding outside of major sheep producing countries like Australia. The pet goat industry in the U.S. is particularly over-looked with regard to paratuberculosis and the infection is sadly very common.
« Previous 1 2 3 4 5 … 18 Next »