JD CONTROL IMPROVES CALF HEALTH & DAIRY INCOME
2020-03-09 18:35:16 A research team in Guelph, Canada demonstrated the significant economic benefit to dairy farms working to control Johne’s disease and neonatal calf diarrhea. The analysis evaluated only changes to farm management (no diagnostic testing). Specifically, they evaluated changes to one or more of the following areas: (1) calf feeding, (2) maternity pen management, and (3) maternity area structure. These two diseases were investigated because control programs for one disease often work to control the other so herd owners get double the benefits when implementing control programs focused on changing farm management methods. This excellent article appeared in the January issue of the Journal of Dairy Science and is Open Access [21 pages with 52 references].
A research team in Guelph, Canada demonstrated the significant economic benefit to dairy farms working to control Johne’s disease and neonatal calf diarrhea. The analysis evaluated only changes to farm management (no diagnostic testing). Specifically, they evaluated changes to one or more of the following areas: (1) calf feeding, (2) maternity pen management, and (3) maternity area structure. These two diseases were investigated because control programs for one disease often work to control the other so herd owners get double the benefits when implementing control programs focused on changing farm management methods. This excellent article appeared in the January issue of the Journal of Dairy Science and is Open Access [21 pages with 52 references].
Abstract
The objective of this study was to perform a cost-benefit analysis (CBA) of a participatory extension model, called Ontario Focus Farms (FF), which was designed to facilitate the adoption of on-farm management practices to control Johne's disease (JD) on Ontario (ON) dairy farms. Partial budget models were developed to estimate the annual herd cost of JD on an average 78-cow Ontario dairy herd and the annual herd cost of neonatal calf diarrhea (NCD). With these estimates, a CBA was developed to assess the simulated net benefits of implementing various on-farm management scenarios (i.e., implementing 1, 2, or 3 of the following: calf feeding, maternity pen management, maternity area structure changes), where the benefits represent a reduction in the annual cost of JD and NCD. These models informed the final CBA assessing the net benefits of FF implementation over a 10-yr period. All monetary values are reported in Canadian dollars (Can$; where 1 Can$ = 0.823 US$ at the time of the study).
The annual herd cost of JD was estimated to be $3,242 ($41.56/cow), and that of NCD was estimated to be $1,390 ($36/heifer calf). When farms were expected to have both JD and NCD, all scenarios, when implemented over a 10-yr period, yielded positive net benefits ranging from $439 to $2,543 per farm when changes to maternity area structure were combined with calf feeding changes. These effects were sensitive to changes in level of disease (JD and NCD) on the farm, and the costs and effects of making changes. The NPV of making any on-farm change when JD was not present on the farm was negative. Overall, FF implementation yielded positive net benefits of $426,351 or $749,808, depending on whether a veterinarian or non-veterinarian served as the facilitator. The NPV was most sensitive to changes in burden of disease, the cost of implementing changes, and the proportion of FF participants that had JD and NCD on the farm. Benefits of FF implementation are also likely to accrue to veterinarians, as a result of professional facilitator training, and the Ontario dairy industry, as a by-product of improved milk quality and safety; therefore, the true net benefits of FF implementation are likely underestimated. Overall, the FF process should be considered an economically viable program and worthy of investment as part of a JD control strategy, as it demonstrates potential to yield positive net benefits for the Ontario dairy industry.

Conclusions from the publication: The total annual herd cost of JD to an average Ontario dairy farm, with a true within-herd prevalence of 10%, was estimated to be $3,242. The total annual herd cost of NCD to an average Ontario dairy farm, with an incidence rate of 23%, was estimated to be $1,390. Implementation of various combinations of calf feeding changes, maternity pen management changes, and maternity area structure changes resulted in varying net benefits in these simulations, depending on the presence of JD and NCD, the true burden of disease, and the specific management practices implemented. Overall, the implementation of the FF process over a 10-yr period yielded positive net benefits, suggesting that its implementation would be valuable for reducing the burden of JD and NCD on Ontario dairy farms.
Comment: As the authors mention, ensuring that producers follow the recommended control program consistently over time is a key factor in the realization of benefits. External farm advisers, such as veterinarians, are critical to this effort. Also, adoption of multiple management changes is most economically beneficial if the farm is dealing with both JD and neonatal calf diarrhea.
JD CASE REPORT: TAMIL NADU, INDIA
2020-03-01 21:29:14A. Karthikeyan and colleagues report on a case of Johne’s disease in the state of Tamil Nadu, India in the International Journal of Current Microbiology and Applied Science (volume 9, issue 1, pages 22-64-2267, 2020).
Abstract
 Johne’s disease (Paratuberculosis), a debilitating chronic granulomatous enteritis of domesticated and wild ruminants that causes huge economic and production losses to the dairy farmers. A six-year-old Jersey crossbred cow was presented with a history of chronic diarrhoea, gradual weight loss, reduction in milk yield and poor response to therapy. Based on history, clinical examination, Ziehl-Neelsen staining and IS900 polymerase chain reaction of the faeces the case was diagnosed as Johne’s disease. It warrants strict implementation of control measures to put off further spread Johne’s disease in dairy animals.
Johne’s disease (Paratuberculosis), a debilitating chronic granulomatous enteritis of domesticated and wild ruminants that causes huge economic and production losses to the dairy farmers. A six-year-old Jersey crossbred cow was presented with a history of chronic diarrhoea, gradual weight loss, reduction in milk yield and poor response to therapy. Based on history, clinical examination, Ziehl-Neelsen staining and IS900 polymerase chain reaction of the faeces the case was diagnosed as Johne’s disease. It warrants strict implementation of control measures to put off further spread Johne’s disease in dairy animals.

Comment: There are more cows in India than in any other country of the world. Cows have a special place in Indian society making Johne’s disease especially difficult to control in cattle.

Read more about sacred cows from Wikipedia: In the Hindu tradition, cows are honored, garlanded and given special feedings at festivals all over India. One is the annual Gopastami festival, dedicated to Krishna and cows.
The cow's nature is represented in Kamadhenu; the goddess who is the mother of all cows. In India, more than 3,000 institutions called Gaushalas care for old and infirm cows. According to animal husbandry statistics there are about 44,900,000 cows in India, the highest in the world. So while some old and infirm cows are treated in Gaushalas, the rest are generally abandoned at public places such as railway stations and bazaars.
Honoring the cow inspires in people the virtues of gentleness and connects them with nature. The cow gives milk and cream, yogurt and cheese, butter and ghee. The milk of a cow is believed to refine a person. The ghee (clarified butter) from the milk is used in ceremonies and in preparing religious food. Cow dung is used as fertilizer, as a fuel and as a disinfectant in homes.
The 16th International Colloquium on Paratuberculosis will be held at Amity University Rajasthan, Jaipur, India, October 17-21, 2022.
IRISH SURVEY FOR MAP IN CATTLE
2020-02-23 22:26:25Research Article
Elvira Ramovic and 9 colleagues reported a pilot study to assess the rate of MAP infection in Irish dairy and beef cattle herds, and much more. The abstract is below and the comments that follow discuss the dairy survey prevalence data specifically. The article is in the Irish Veterinary Journal and is Open Access.
Abstract
Background: Dairy and beef cattle can be reservoirs of many pathogens, including Salmonella and Mycobacterium avium subsp. paratuberculosis (MAP), the causative agent of Johne’s disease (JD). Farm environments may provide potential entry points for the transmission of infectious agents into the food chain. Antibiotics are used to treat a wide variety of infections on farms, and administration of antimicrobial agents to cattle is considered to be a driving factor for antimicrobial resistance (AMR). Control of JD and AMR are priority for animal health initiatives in Ireland. A national JD pilot programme was introduced by Animal Health Ireland in 2014, while the national action plan launched by Department of Health and Department of Agriculture, Food and Marine introduced in 2017 aims to improve the surveillance of AMR. The current investigation was undertaken as a pilot study to determine the proportion of herds positive for MAP, Salmonella species (Salmonella spp), commensal Escherichia coli (E. coli), Extended-spectrum beta-lactamase (ESBL) AmpC β-lactamase and carbapenemase-producing E. coli from 157 environmental faecal samples in Irish farms.
Results: MAP was detected in 10.2% of samples collected; on culture in 4 (4.9%) of the dairy herds and from 1 (1.3%) of the beef/suckler herds, and by PCR in 10 (12.3%) and 6 (7.9%) of these herds respectively. All culture positive herds were also positive by PCR. An additional 11 herds were positive by PCR only. Salmonella was not detected, while commensal E. coli were isolated from 70.7% of the samples (111/157) with 101 of these isolates shown to be fully susceptible to all antimicrobials tested. Of the 27 presumptive ESBL AmpC β-lactamase producing E. coli detected, one isolate was resistant to ten antimicrobials, nine isolates were resistant to nine antimicrobials, and four isolates were resistant to eight antimicrobials. Carbapenemase-producing E. coli were not isolated.
Conclusions: The results highlight the importance of monitoring farm environments for Johne’s disease. This disease is a growing concern for dairy and beef producers in Ireland, and sampling the farm environment may offer a useful means to rapidly screen for the presence of MAP. Non-pathogenic common enteric commensal and multiple-drug-resistant E. coli may contribute to AMR acting as a reservoir and transferring resistance to other species/pathogens in the environment.
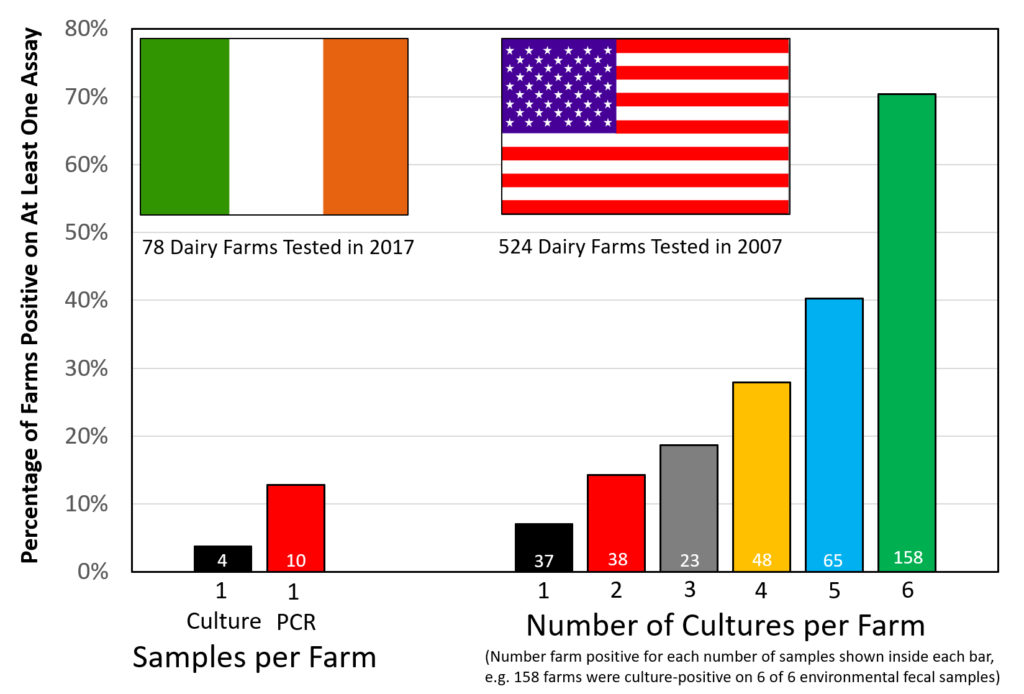
Comment: As they say, “the devil is in the details”.
The above graphic contrasts the two similar surveys for MAP on dairy farms, both done using environmental fecal samples.
The Irish survey used culture methods on the first sample finding 4 farms positive of the 78 sampled (5.1%). The second sample was tested using IS900 PCR finding 10 of 78 (12.8%) positive. All 4 culture-positive farms were also PCR-positive. Were more farms detected because PCR is more sensitive, or because a second sample increases the chances of detecting an infected farm?
The US survey only used culture methods to judge the farm’s MAP infection status but collected 6 samples per farm. The US data indicates that: the more you look the more you find. Over 70% of US dairy farms had a least one culture-positive sample. From this Lombard et al. estimated the true herd-level MAP infection ate in US dairy herds to be 91.1%.
Clearly there are more MAP-infected farms in the U.S. than in Ireland, however, it is tricky to estimate just how many would have been detected if only 2 samples were tested, i.e. which 2 of the 6 tested samples would you count? I will leave that for the modelling experts. For the purposes of this news item it is not that important.
However, since this was a pilot study, it seems that a larger study is warranted that: 1) randomly selects farms, 2) uses PCR as the probably more sensitive and definitely more cost-effective diagnostic test, 3) measures the analytical sensitivity of the test based on spiked samples to better understand the accuracy of the measurement tool, and 4) collects more than 2 environmental fecal samples per farm.
For detection of MAP-infected dairy farms with a low infection prevalence, e.g. 0 to 12%, as in the Irish situation, Lombard et al. estimated the herd-level sensitivity of a single environmental fecal culture at 33.3% and two environmental fecal cultures at 44.4% (see Table 1 in Lombard, 2013). If Lombard et al. are correct, the Irish survey may have under-estimated the true MAP infection status of its dairy farms.
BUT to be fair - it was only a pilot study! (and nobody asked my opinion). :-)
MAP RISK FACTORS IN CHILEAN DAIRY HERDS
2020-02-17 21:16:45Research Article
Cristobal Verdugo, Maria Francisca Valdes, and Miguel Salgado from the Instituto de Medicina Preventiva Veterinaria, Facultad de Ciencias Veterinarias, Universidad Austral de Chile, Valdivia, Chile described herd level risk factors for Mycobacterium avium subsp. paratuberculosis (MAP) infection and clinical incidence in dairy herds in Chile. Their publication will appear in Preventive Veterinary Medicine, volume 176, March 2020 and is available online now (6 pages with 56 references). Unfortunately, it is not Open Access.

Holstein with clinical Johne's disease photographed in Chile by M.T. Collins.
Abstract
The study objective was to identify risk factors associated to: i) the infection by Mycobacterium avium subsp. paratuberculosis (MAP), and ii) paratuberculosis clinical incidence in Chilean dairy herds. A random sample of forty herds with previous history of MAP infection was selected. At herd level, all lactating cows were tested using a commercial ELISA kit. On the sampling date, a questionnaire gathering information on herd demographics, husbandry practices, and biosecurity measures was applied. Additionally, the farm manager/owner was surveyed regarding the number of paratuberculosis compatible clinical cases (CCC) in the last 12 months. Two Bayesian generalized linear mixed effect models were used to evaluate the association between the questionnaire data, and the proportion of truly infected animals (model 1) or the number of CCC (model 2).
A total of 4963 animals were sampled with an average apparent prevalence of 6.3 % (95 % confidence interval (4.0–8.0%). All sampled herds presented seropositive animals. Forty eight percent of the herds did not observe any CCC in the last year. Although, among those herd that did report CCC, a median of two cases per year was estimated. Model outputs showed that the proportion of truly infected animals and CCC reporting rates are associated to management practices. Specifically, positive associations were observed for feeding of calves exclusively with milk replacer, and the distance between the milking parlor and the calves’ barn. Additionally, CCC reporting rates were higher in farms that recently purchased animals, and where the distance between the milking parlor and the calves’ barn was less than 30 m.
Author’s Conclusions : “…the present research was able to find herd level factors associated to an increase in the risk of infection and the chances of reporting CCC. The reported factors put an emphasis on the need of introducing or improving basic biosecurity measures, especially in relation to the hygiene of calves, limiting the contact of young animals with manure from adult cows. The investment in such measures would not only be beneficial for MAP control but would improve the general health of the herd, possibly reducing the use of veterinary drugs.
Comment: The association of MAP infections with feeding calves milk replacer should be interpreted with caution. While milk replacer has been shown to harbor viable MAP (Grant 2017), it is also possible that preparation of the milk replacer on farms allowed fecal contamination which carried MAP into the re-hydrated product before feeding.
ON-FARM PASTEURIZER VS MAP
2020-02-10 20:28:23Research Article
K. Fechner and colleagues from the Department of Animal Sciences, Faculty of Agricultural Sciences, University of Göttingen reported about on-farm HTST (high-temp. short-time) pasteurizer's ability to kill MAP in the December 2019 issue of the Journal of Dairy Science. [not Open Access]
[caption id="attachment_3192" align="aligncenter" width="640"] HTST pasteurizer on a farm in Wisconsin[/caption]
HTST pasteurizer on a farm in Wisconsin[/caption]
Abstract
Feeding pasteurized milk to suckling calves is a popular practice used increasingly on dairy farms. Waste milk is frequently fed to calves because of its high nutritional value and economic benefits compared to milk replacement products. However, one of the disadvantages of feeding waste milk is the potential for exposure to a high number of bacterial contaminants, which may lead to serious illnesses or infections in calves. One of these contaminants is Mycobacterium avium ssp. paratuberculosis (MAP), the causative agent of Johne's disease (paratuberculosis). The transmission and distribution of paratuberculosis in dairy herds occurs mostly through the feeding newborn calves with contaminated colostrum or milk, because this age group is believed to be most susceptible to infection. To reduce the risk of transmission of pathogens, on-farm pasteurization of milk has become increasingly popular.
In this study, we analyzed the efficacy of a new commercial high-temperature, short-time pasteurizer (73.5°C for 20 to 25 s) in terms of MAP inactivation under experimental on-farm conditions. The pasteurizer uses a newly developed steam-heating technique, allowing for the pasteurization of the transition milk without clumping. In 3 independent trials, we spiked fresh raw milk samples to a level of 107 or 104 viable MAP cells/mL before pasteurization. We examined the thermal inactivation and viability of MAP using culture and a D29 bacteriophage-based assay. To verify the identity and number of MAP cells, we also performed PCR assays.
Pasteurization of the inoculated milk (107 and 104 MAP cells/mL) resulted in a remarkable reduction in viable MAP cells. The mean inactivation rate of MAP ranged from 0.82 to 2.65 log10 plaque-forming units/mL, depending on the initial MAP amount inoculated and the addition of conservative agents to the pasteurized milk. Nevertheless, approximately 103 MAP cells/mL remained viable and could be transferred to calves after high-temperature, short-time pasteurization of milk.
[caption id="attachment_3193" align="aligncenter" width="640"] Milk: an effective way to transmit Johne's disease.[/caption]
Milk: an effective way to transmit Johne's disease.[/caption]
Comment: Yet again, research has shown that HTST pasteurization [minimum: 161F (72C) x 15 sec] fails to reliably kill 100% of MAP bacteria. And, this study used a slightly higher temperature and longer time [73.5°C for 20 to 25 s]. Achieving 100% kill of MAP by pasteurization is primarily a function of temperature AND time AND the number of MAP in the raw milk.
Recent research by P. Steuer and colleagues in Chile reported finding up to 106 MAP/mL raw milk on Chilean dairy farms with ELISA-positive animals (see P. Steur et al., Is the transmission of Mycobacterium avium subspecies paratuberculosis (MAP) infection through milk intended to feed calves an overlooked item in paratuberculosis control programs?, Tropical Animal Health and Production (vol 52 issue 1, pp 89-94, January 2020).
Many explanations for the heat-resistance of MAP have been suggested, e.g. formation of clusters of bacterial cells (up to 100) hindering heat penetration, location inside host cells (protected), and perhaps most importantly the ability to form tenacious spore-like structures. For more on spore formation see Lamont et al. PLOS ONE, January 24, 2012.)
DAIRY: IMPACT OF DAM’S JD STATUS
2020-02-03 19:24:05Research Article
S. Patterson and colleagues in the UK reported on the impact of a dairy cow’s Johne’s ELISA status on the likelihood her calf will also be MAP-infected (ELISA-positive). Their study is reported in the journal Preventive Veterinary Medicine (in press, corrected proof, available online – not Open Access).
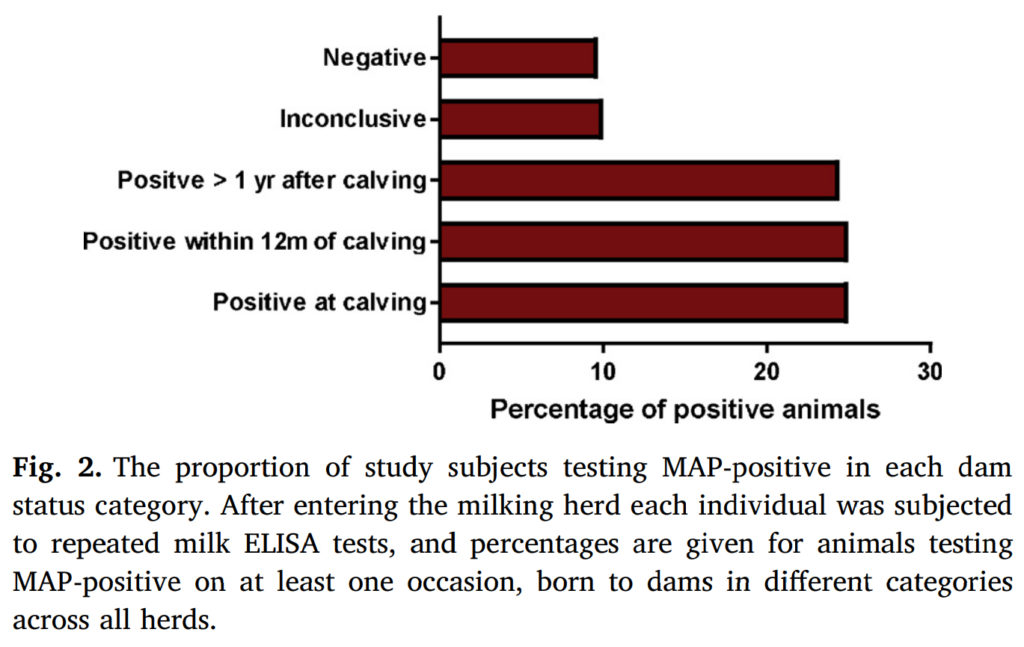
Abstract
Johne’s disease, caused by Mycobacterium avium subsp. paratuberculosis (MAP), is a chronic condition of dairy cattle, and is endemic in the UK. Lack of understanding of the relative importance of different transmission routes reduces the impact of control scheme recommendations.
The long incubation period for Johne’s disease makes evaluation of control schemes difficult, and so this long-term cohort study offers a rare and valuable insight into the disease epidemiology. A longitudinal study was carried out following a cohort of 440 UK dairy cows in 6 herds recruited in 2012-2013. Individuals entering the milking herd were routinely monitored for the presence of MAP using quarterly milk ELISA testing. Using a Cox proportional-hazards regression model the relationship between time until first detection of infection and dam MAP status was investigated. We then compared the magnitude of the effect of dam status with that of other risk factors in order to understand its relative importance.
Dam status was found to be the only observed factor that was significantly associated with time to an individual testing MAP-positive (p = 0.012). When compared to negative dams, we found a marginally significant effect of having a positive dam at time of calving, that increased the hazard of an individual testing positive by a factor of 2.6 (95% confidence interval: 0.89–7.79, p = 0.081). Further positive associations were found with dams becoming positive after the birth of the subject; a dam seroconverting within 12 months post parturition being associated with a 3.6 fold increase in hazard (95% confidence interval: 1.32–9.77, p = 0.013), and dams seroconverting more than a year after calving increased the hazard by a factor of 2.8 (95% confidence interval: 1.39–5.76, p = 0.004).
These results suggest that cows may be transmitting MAP to their offspring at an earlier stage than had previously been thought, and so raise important questions about how this transmission may be occurring. The results of the study may have important practical implications for the management on-farm of the offspring of MAP-positive animals, with the potential to vastly reduce the time required to eliminate this chronic disease.
Comment: For faster control of Johne’s disease in dairy herds it is advisable to tell herd owners not to retain as herd replacements any heifer born to a cow that tests positive for Johne’s disease. This study supports that recommendation (using ELISA as the Johne’s disease test) and extends it to include calves born to cows that test positive AFTER calving. Not culling calves born to test-positive dams will result in more MAP-infected heifers entering the herd as replacements and then infecting more calves thereby slowing progress of a control program. That said, in my experience it is difficult enough to get herd owners to act on positive test results on cows, much less act on the probabilities that their calves are also infected. Hopefully, this report and others like it will help to strengthen culling recommendations to include calves born to ELISA-positive dams resulting in more successful control of Johne’s disease in dairy herds.
MAP-INFECTED CARNIVORES IN PORTUGAL
2020-01-26 18:11:07 Today's news: Another report of probable MAP “spillover” from domestic animals to wildlife. Monica Cunha from the National Institute for Agrarian and Veterinary Research, Oeiras, Portugal and colleagues report detection of MAP by PCR in 10% red fox (Vulpes vulpes), 6% Egyptian mongoose (Herpestes ichneumon) as well as stone marten (Martes foina) and common genet (Genetta genetta) – a first time report of MAP in genet. This Open Access publication appeared January 21 in Nature.com, Scientific Reports. [14 pages with 88 references]
Today's news: Another report of probable MAP “spillover” from domestic animals to wildlife. Monica Cunha from the National Institute for Agrarian and Veterinary Research, Oeiras, Portugal and colleagues report detection of MAP by PCR in 10% red fox (Vulpes vulpes), 6% Egyptian mongoose (Herpestes ichneumon) as well as stone marten (Martes foina) and common genet (Genetta genetta) – a first time report of MAP in genet. This Open Access publication appeared January 21 in Nature.com, Scientific Reports. [14 pages with 88 references]

Common genet photo from Wikipedia.
Abstract
Mycobacterium avium subsp. paratuberculosis (MAP) is the etiological agent of Johne’s disease or paratuberculosis, a chronic infection affecting domestic ruminants worldwide. Despite sporadic reports of MAP occurrence in non-ruminants, information on the risk factors predisposing for infection is still scarce and evidence of transmission paths linking the livestock-wildlife-environment interfaces also remains lacking. In this study, we predicted that environmental, host-related, land use and human driven disturbance factors would modulate carnivore exposure to MAP. To test these hypotheses, we performed a retrospective survey, based on microbiological and molecular methods, in mainland Portugal including five sympatric species from the Herpestidae, Canidae, Viverridae, and Mustelidae families (n = 202) and examined 16 variables as putative predictors of MAP occurrence. Molecular evidence of MAP using IS900 as proxy was demonstrated in 7.43% (95%CI: 4.55–11.9) of surveyed carnivores, the highest proportions being registered for red fox (Vulpes vulpes) (10%; 95%CI: 4.0–23) and Egyptian mongoose (Herpestes ichneumon) (6.0%; 95%CI: 3.2–11). We demonstrate that important species of the Mediterranean carnivore guild, such as stone marten (Martes foina) and common genet (Genetta genetta), may also be exposed to MAP, being this the first time that occurrence in genet is reported. The high proportion of DNA-positive specimens, concurrent with the apparent lack of gastro-enteric lesions and molecular confirmation of IS900 in feces, argue for the presence of subclinical carriers that occasionally shed bacteria, potentially aiding as source of infection to susceptible species and possibly contributing for environmental contamination. Achievement of MAP isolation would prove beyond any doubt that MAP is present in this wildlife population. Ecological modelling results suggested that the probability of MAP infection using IS900 as proxy in mongoose is positively associated with higher altitude and temperature stability, as well as with lower annual rainfall. Density of livestock farms was found not to be a significant predictor, which may indicate that the livestock-wildlife interface is probably not important as an infection route for mongoose.

Egyptian mongoose photo from Wikipedia.
Concluding remarks excerpt: Suspicion for the ecological spread of MAP reinforces the need for increased surveillance and action. Quantitative information on the prevalence and concentration of MAP in putative contaminated sources of exposure is required.
Comment: This report adds to the growing body of literature about MAP spread to wildlife. It is critical to establish whether the transmission of MAP is a “two-way street”, with MAP going back and forth between domestic animals and wildlife, or instead that wildlife are a “dead-end host”. i.e. transmission only from domestic animals to wildlife ("one-way street"). The answers will likely depend on the type of wildlife, i.e. wild ruminants probably develop permissive infections shedding MAP in ways that can then infect domestic animals, while wild carnivores may be dead-end hosts. Read more about non-ruminants that have been found to be MAP-infected on this website page.
DAIRY GOATS THREATENED BY JOHNE’S
2020-01-20 19:41:18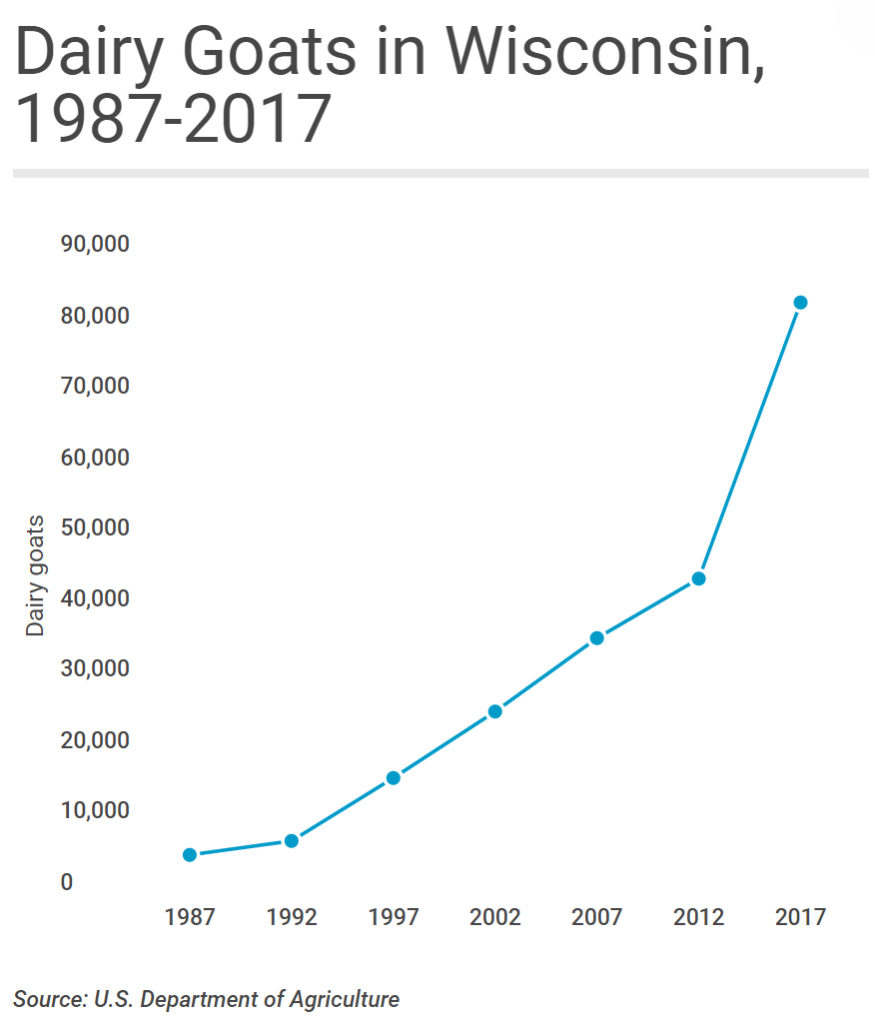
As reported in the Wisconsin State Farmer, September 16, 2019:
Wisconsin's self-proclaimed moniker as "America's Dairyland" is taking on fresh meaning in the 21st century thanks to a growing market for milk from an animal that bleats rather than moos. While the state's traditional dairy cattle industry continues to hemorrhage producers at a record pace, Wisconsin's dairy goat industry is in the midst of a long-term, and accelerating, growth spurt. Indeed, in 2019 Wisconsin can reasonably claim to be America's dairy (goat) land.
Data from the United States Department of Agriculture, which counts livestock across the United States every 5 years, show just how much Wisconsin dominates the nation's dairy goat industry. In 2017, the most recent year the USDA surveyed producers, the size of Wisconsin's dairy goat herd easily topped the nation at more than 83,000-head. California came in a distant second, with some 43,000 dairy goats, while Iowa, Texas and Missouri rounded out the top five.
It's not only the sheer size of Wisconsin's dairy goat herd that stands out: The state also leads the nation in the value of sales from dairy goat operations and is the epicenter of national growth in goat dairy.
Read the rest of this excellent article that contains many useful links here.
Comment:
Rapidly expanding animal industries commonly overlook biosecurity risks feeling compelled to buy animals regardless of health status. Many of these new dairy goat operations have 5,000 to 10,000 goats. Operations only can attain this size by purchase of goats from many different herds. Purchasing animals is the #1 way by which herds, whether of cattle, sheep or goats, become infected with MAP. This then leads to long-term problems of animal health, animal welfare, animal productivity and animal longevity which all translate to a negative economic impact due to Johne’s disease.
Johne’s disease is common among all goat breeds.
- France found that 55.2% of dairy goat herds had one or more ELISA-positive goats.
- In Cyprus 50% of dairy goat herds were seropositive.
- In Ontario in Canada 83% (estimated true prevalence) of dairy goat herds were MAP-infected.
- In Spain 87.5% of dairy goat herds were seropositive.
- I found no such surveys in U.S. dairy goats but a study of Boer goats in Missouri reported that an estimated 54.7% of Boer goat herds are seropositive for Johne’s disease.
Many owners fail to realize their goats are MAP-infected and therefore do not implement control measures with the result that the infection continues spreading. In this article in the Wisconsin State Farmer, I also observed that links provided in this article to goat associations and experts never once mention biosecurity concerns or the threat of Johne’s disease. So unfortunate!
Goat owners and veterinarians must be much more aware of Johne’s disease and implement biosecurity protocols and on-farm control measures to mitigate the damage caused by Johne’s disease. When MAP becomes recognized as a food-borne zoonotic pathogen, these large goat herds with face a daunting task to control or eradicate this infection.
The age-old wisdom that “prevention pays” should be heeded.
MAP THREATENS ENDANGERED DEER
2020-01-11 19:57:31A rare and beautiful sight, the Southern Patagonian huemul deer (Hippocamelus bisulcus) is an icon of Patagonia Park and of Chile. A native species, the huemul is Chile’s national animal, found on the country’s coat of arms, alongside the Andean condor. Despite its high level of recognition, the huemul is classified as endangered by the IUCN, with a global population of less than 1,500 individuals. With 10% of these remaining deer residing within Patagonia Park’s boundaries, Conservacion Patagonica has made huemul deer recovery the cornerstone of its wildlife program.

Paulo Corti and colleagues from Universidad Austral de Chile, Valdivia, Chile describe recovery of MAP from 3 of three fecal samples from huemul deer in Patagonia in The Australian Journal of Veterinary Science (52:33-35, 2020).
Abstract
In a huemul (Hippocamelus bisulcus) population sympatric with cattle, we found evidence of Mycobacterium avium subsp. paratuberculosis (MAP) infection. Three huemul faecal pellet samples and two cow pats were collected and cultured for MAP presence. DNA was then extracted for PCR analysis of all signal-positive cultures. To assess whether MAP isolates obtained from huemul faeces were associated with typical MAP isolated from livestock, positive confirmed culture samples were sub-typed using a combination of five Mycobacterial Interspersed Repetitive Unit-Variable Number Tandem Repeat Analysis and one Short Sequence Repeat analysis markers. All faecal samples from both species were MAP positive. One huemul presented a different bacteria profile genotype not described before, suggesting that huemul and cattle in Patagonia could carry a unique MAP strain.
Comment: The primary reservoir for MAP is in domesticated ruminants such as dairy cattle, beef cattle, sheep and goats. However, MAP infections in these livestock readily spread to wild ruminants and this first-time report of finding MAP in huemel deer illustrates the concept of infection “spillover”.
 Spillover is the title of an excellent book by David Quamenn which focuses on the spillover of infectious diseases from animals to humans, so called zoonotic infections. MAP infections clearly spillover to wild ruminants, and non-ruminants, including humans. This website provides more details about MAP infections of dairy cattle, beef cattle, goats, sheep, deer & elk, bison, water buffalo, wild ruminants, zoo ruminants, and non-ruminants. A separate page is devoted to the zoonotic potential of MAP.
Spillover is the title of an excellent book by David Quamenn which focuses on the spillover of infectious diseases from animals to humans, so called zoonotic infections. MAP infections clearly spillover to wild ruminants, and non-ruminants, including humans. This website provides more details about MAP infections of dairy cattle, beef cattle, goats, sheep, deer & elk, bison, water buffalo, wild ruminants, zoo ruminants, and non-ruminants. A separate page is devoted to the zoonotic potential of MAP.
MAP is a promiscuous, insidious, zoonotic, foodborne pathogen that threatens the economic viability of livestock producers, the health and well-being of wildlife, zoological collections of wild ruminants (many of which are endangered), and humans. It deserves far more research funding and control program investment than it currently receives.
SHEEP AND SAMPLE POOLING
2020-01-05 17:33:52Research Report
Mathevon and colleagues from Ecole Nationale Vétérinaire de Toulouse, Toulouse Cedex, France reported on optimal sheep sample pooling strategies for serum samples (for ELISA testing) or fecal samples (for qPCR testing). Their report appeared December 26, 2019 in PLoS ONE and is OPEN ACCESS. [18 pages with 47 references]
Abstract
The aim of our study was to evaluate the flock sensitivity and specificity of fecal qPCR and serum ELISA using pooled samples for screening paratuberculosis in French sheep.
Using individual feces with low or high qPCR Ct values from ewes sampled in 14 infected flocks, a total of 555 pools of size 5, 10 and 20 were created by diluting individual materials in negative feces and analysed using a commercial IS900 qPCR kit. The relative performances of pooled serum ELISA analysis were evaluated based on the analysis of 181 different pools of size 5 and 10, composed of individual serum samples of various individual S/P values. Results showed that for pools of size 5, 10 or 20, individual fecal samples with low Ct values were invariably detected. Conversely fecal samples with high Ct values were associated with a lower detection rate in both pools of size 5 (87.0% to 90.0%), 10 (63.0% to 70.7%) and 20 (46.7% to 60.0%). After lowering the decision threshold to 25% and 15% for serum pools of size 5 and 10 respectively, the pooled serum ELISA relative sensitivity ranged between 62.2% and 100.0% depending on the composition of the pools.
Finally, a simulation study was carried out to evaluate the performances of 16 screening strategies at flock level, with varying pool size (5 to 20) and number (5 to 60). The use of pooled serum ELISA led to very false positive detection rate ranging between 37.6% and 91.8% in paratuberculosis free flocks and prevents its further use in that context. For infection prevalence ≤ 5%, the flock sensitivity based on pooled fecal qPCR ranged between 39.0% (5 pools of size 10) and 99.9% (300 sampled individuals, with pools of size 5,10 or20), and was always above 93% when the infection prevalence was greater or equal to 15%. We conclude that pooled-fecal qPCR but not pooled-serum ELISA could be a useful tool to detect sheep flocks infected with paratuberculosis.
Comment: The authors focused their discussion on diagnostic tests useful for screening of sheep flocks for ovine Johne’s disease (OJD) where the primary goal is to detect MAP-infected flocks, i.e. maximize “flock sensitivity” and “flock specificity”, at the least cost. This should not be confused with the use of fecal pooling strategies for qPCR testing in known infected flocks where the goal is to identify individuals in the flock that should be culled or isolated.
In MAP-infected flocks there are two equally important goals: 1) to limit the cost of testing the whole flock, and 2) to limit the cost of testing the individual samples, i.e. to have the fewest number of qPCR-positive pools. This second goal is important because each of the fecal samples comprising a qPCR-positive pool must be individually tested by qPCR in order to identify each MAP-infected sheep adding significant costs, e.g. roughly $30 x 5 = $150 per PCR-positive pool (based on WVDL testing rates).
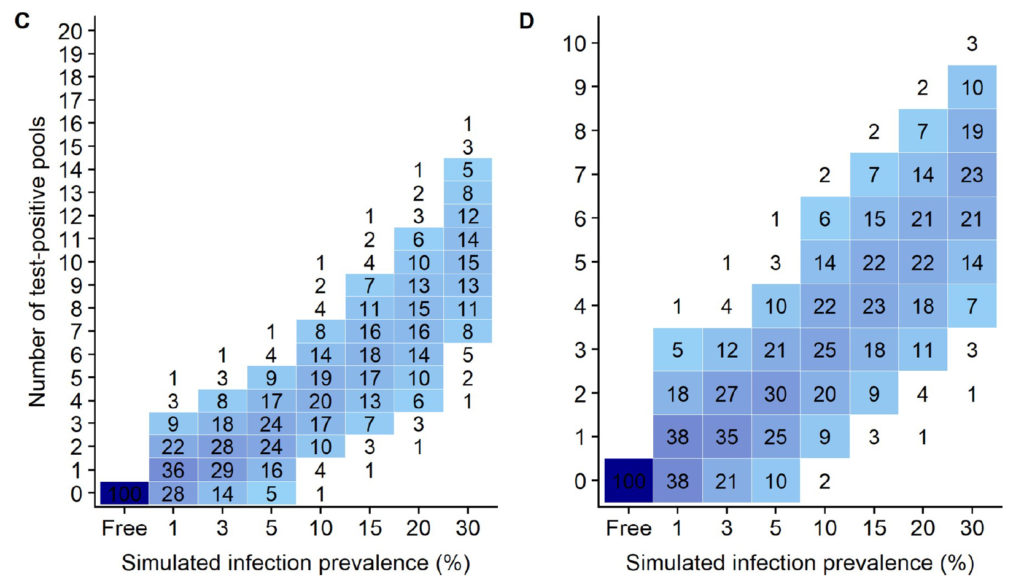
Using the simulation data from this publication (shown above), for flocks with a MAP infection prevalence of 10%, the simulation data (Fig 4 C & D in the publication as shown above) indicate that using 20 pools of 5 fecals/pool, 93% of flocks would have 7 or fewer positive pools which would then cost an additional $1,050 to test each fecal individually by qPCR (7 pools x 5 fecals/pool x $30/qPCR = $1,050). By contrast, if these same flocks used 10 pools of 10 fecals/pool, then 92% of flocks would have 5 or fewer qPCR-positive pools resulting in a cost of $1,500 to individually test the fecal samples making up those pools (5 pools x 10 fecals/pool x $30 = $1,500).
The cost of testing by pooled PCR at the WVDL is $34/pool so would cost $340 for 10 pools of 10 fecals/pool or $680 for 20 pools of 5 fecals/pool. Thus, while testing 20 pools (5 fecals/pool) would initially cost $340 more, the cost of testing the individual fecals comprising each qPCR-positive pool does not offset the cost of testing fewer pools in this example.
These findings are based on a simulation of random pooling of fecal samples described in the publication. As mentioned in a previous news posting, strategic pooling by animal age can help limit the number of test-positive pools Kalis (2000). This is because animals of similar age experienced similar risks of becoming MAP-infected when young. Thus, age-based pooling (strategic) results in more MAP-infected animals likely to be in the same pool and conversely the fewest number of positive pools for the flock or herd as a whole.
You can read more about a clustering of MAP-infections by age and season in the publication by Zare et al. titled: Evidence of birth seasonality and clustering of Mycobacterium avium subspecies paratuberculosis infection in US dairy herds. Preventive Veterinary Medicine 112: 276– 284, 2013. However, the clustering effect may not be as pronounced for sheep which do not have lambs thought the year.
In closing, this discussion highlights the importance of utility analysis when judging the merits of diagnostic tests for MAP. For more on this topic see Garder et al. Consensus-based reporting standards for diagnostic test accuracy studies for paratuberculosis in ruminants. Preventive Veterinary Medicine 101: 18-34, 2011.
« Previous 1 … 9 10 11 12 13 … 18 Next »